Abstract
Aims: Patients with multiple tumour localisations pose a particular problem to the pathologist when the traditional combination of clinical data, morphology, and immunohistochemistry does not provide conclusive evidence to differentiate between metastasis or second primary, or does not identify the primary location in cases of metastases and two primary tumours. Because this is crucial to decide on further treatment, molecular techniques are increasingly being used as ancillary tools.
Methods: The value of comparative genomic hybridisation (CGH) to differentiate between metastasis and second primary, or to identify the primary location in cases of metastases and two primary tumours was studied in seven patients. CGH is a cytogenetic technique that allows the analysis of genome wide amplifications, gains, and losses (deletions) in a tumour within a single experiment. The patterns of these chromosomal aberrations at the different tumour localisations were compared.
Results: In all seven cases, CGH patterns of gains and losses supported the differentiation between metastasis and second primary, or the identification of the primary location in cases of metastases and two primary tumours.
Conclusion: The results illustrate the diagnostic value of CGH in patients with multiple tumours.
Keywords: comparative genomic hybridisation, pathology, diagnostic consults, multiple tumours
In day to day routine work, pathologists are faced with cases that take more than a standard haematoxylin and eosin stain to solve the diagnostic problem and answer the clinician’s questions. Patients with multiple tumour localisations belong to a special category in this respect. Of course, in a large number of cases, in patients with a known and well characterised primary tumour, the second tumour can be confirmed—at least for all practical purposes—to be a metastasis of this primary. In fact, this decision in general is not only based on the histological comparison between the two tumour samples, but also on the clinical plausibility. For example, adenocarcinoma in the liver is regarded as a metastasis of the patient’s colorectal cancer, the squamous cell cancer in a jugular lymph node as a metastasis of the patient’s laryngeal cancer, and adenocarcinoma in an axillary lymph node as a metastasis of the patient’s breast cancer. In other cases, the situation is less obvious, and the question of whether the patient has a metastasis of his primary cancer, or whether a second primary cancer has occurred, is difficult to answer, even though this may be very important for clinical decision making. In these cases, the question is: should the patient be regarded as suffering from (high stage) metastatic disease, or has a low stage second primary occurred, and should the patient be treated accordingly with an intention to cure? The pathologist plays a key role in this process of decision making and is expected to produce the ultimate answer. However, it is obvious that this is not always an easy task, and frequently additional techniques are required. Immunohistochemistry is often the initial method used, but is frequently not enough, either because certain immunophenotypes are common to several types of cancers, or because tumours are negative for almost all markers. In addition, different samples from the same tumour may show extreme phenotypical and immunohistochemical heterogeneity. Therefore, it seems logical to focus not only on the phenotype, but also to look at the genotype. In our study, we present several cases in which comparative genomic hybridisation (CGH) was applied in an attempt to reach a diagnosis.
CGH is a technique that allows the detection of chromosomal copy number changes without the need for cell culturing.1–3 It gives a global overview of chromosomal gains and losses throughout the whole genome of a tumour. Thus, CGH is a relatively fast screening technique that can point at specific chromosomal regions that have been altered in the genome. Because no cell culturing is required for CGH, this technique has enabled tremendous progress in the analysis of chromosomal changes in solid tumours. Applications of CGH in cancer research include screening of tumours for genetic aberrations4,16 (see also http://www.helsinki.fi/~lgl_www/CMG.html), searching for genes involved in the carcinogenesis of particular subsets of cancers,17 analysing tumours in experimental models to provide more insight into tumour progression,17,18 diagnostic classification,17 and prognosis assessment.17–20 In addition to these oncological applications, CGH analysis has also been used to study chromosomal aberrations in fetal and neonatal genomes.21,22
“Comparative genomic hybridisation gives a global overview of chromosomal gains and losses throughout the whole genome of a tumour”
CGH may prove to be useful for the differentiation between a metastasis or second primary, and the identification of a primary location in cases of metastases and two (or more) primary tumours. To investigate this possibility, we decided to analyse several cases with CGH and to look for the concordance of the chromosomal aberrations between multiple tumour samples within one patient.
METHODS
DNA was isolated from paraffin wax embedded, formaldehyde fixed tissue according the protocol described previously.3 CGH was performed as described previously,3 and the results were analysed with an Applied Imaging workstation (Applied Imaging, Newcastle upon Tyne, UK). In total, 18 tumours from seven patients were analysed by CGH.
Results
Figure 1 summarises the results.
Figure 1.
Summary of the comparative genomic hybridisation results for all seven patients. SCC, squamous cell carcinoma.
Patient 1
A 66 year old woman presented with a polypoid tumour of the gastric cardia. After resection, histopathological examination showed a focus of poorly differentiated adenocarcinoma arising in a villous adenoma with superficial invasion of the submucosa. Surgical resection margins and a total of five regional lymph nodes were free of tumour. After two years, on endoscopical examination, a flat lesion was seen in the squamous epithelium of the distal oesophagus within 5 cm of the surgical anastomosis. Histopathology of the biopsy and the subsequent mucosectomy specimen showed a superficially invasive squamous cell carcinoma. Although the two tumour samples showed a different histology, given the close topographical relation between both lesions, the clinicians were seeking for confirmation that the oesophageal tumour was a second primary. Two samples of the gastric tumour (the villous and the solid part) and one sample of the oesophageal tumour were analysed by CGH. The squamous cell carcinoma showed 15 chromosomal aberrations, whereas the two samples of the gastric tumour showed only three and four chromosomal aberrations, respectively. In addition, the pattern of aberrations of the squamous carcinoma differed from the two patterns seen in the adenocarcinoma. These findings are in agreement with the observations that, in general, squamous cell carcinomas show a more complex pattern of chromosomal changes than adenocarcinomas.12 The CGH results showed that the tumour in the gastric cardia and the tumour in the distal oesophagus were genetically unrelated and confirmed that the oesophageal tumour was a second primary, so that there were no indications of metastasis of the primary tumour.
Patient 2
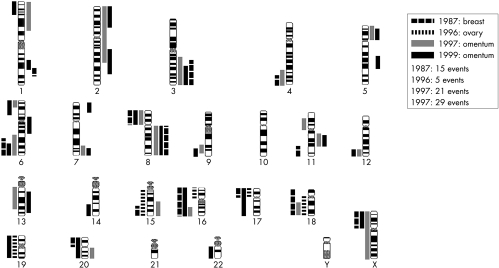
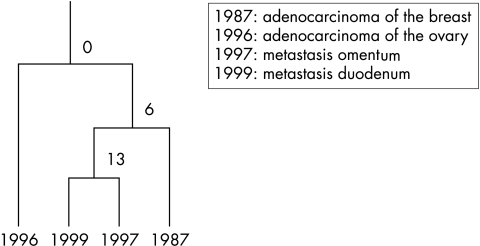
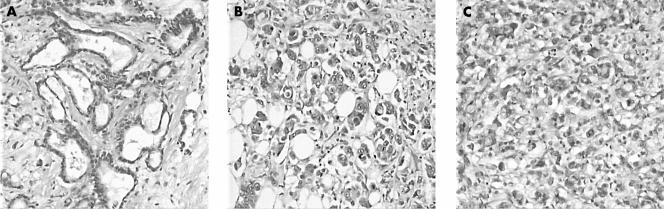
A 51 year old woman presented with a medullary type of breast cancer that was locally radically resected. There were no lymph node metastases. Nine years later she presented with an 11 cm poorly differentiated adenocarcinoma of the left ovary and a 1.5 cm tumour with similar histology in the right ovary; no abdominal metastases were present. Another year later, she had a metastatic tumour removed from the omentum, which proved to be a poorly differentiated adenocarcinoma. Two years after that she underwent partial duodenectomy because of an obstructing tumour mass, which on microscopic examination also proved to be a poorly differentiated adenocarcinoma. Clinically, the ovarian tumours were thought to represent a second primary, although histologically discrimination between the tumours (of the breast, ovary, duodenum, and omentum) was not completely straightforward (fig 2). BRST2 immunohistochemistry was positive in the breast tumour and the metastases in the omentum and duodenum. The two metastases had 13 CGH events in common (fig 3), and therefore were thought to have developed from the same primary tumour. The breast carcinoma from 1987 shared six events with the metastases, whereas none of these six occurred in the ovarian tumour from 1996 (fig 4). It was concluded that the two metastases were derived from the breast carcinoma and that the ovarian cancer was a second primary.
Figure 2.
Histology of the four tumours in patient 2. (A) Breast lesion (1987); (B) tumour in the omentum (1997); (C) ovarian tumour (1996); (D) tumour in the duodenum (1999). All tumours showed solid fields of tumour cells with the occasional glandular lumen.
Figure 3.
Overview of comparative genomic hybridisation events in the four lesions described in patient 2. The bars on the right of each ideogram represent gains, and the bars on the left losses.
Figure 4.
Summary of the results from patient 2. The two metastases have 13 comparative genomic hybridisation events in common and are therefore considered to have developed from the same primary tumour. The breast carcinoma from 1987 shared six events with the metastases, whereas none of these six occurred in the ovarian tumour from 1996. It was concluded that the two metastases were derived from the breast carcinoma and that the ovarian cancer was a second primary.
Patient 3
A 76 year old woman underwent a lumpectomy for a 3 cm large ductal adenocarcinoma in the right breast. The resection margins were free but the sentinel node showed a metastasis. Subsequent lymph node dissections of the right axilla yielded nine lymph nodes free of tumour. Three years later the patient presented with an undifferentiated carcinoma in the bladder. On immunohistochemical evaluation, both tumours were positive for cytokeratin, CAM5.2, AE1/3, and the progesterone receptor. In addition, the breast cancer was positive for the oestrogen receptor (bladder negative) and the bladder was positive for cytokeratin 7 (CK7) and CK20 (breast negative). Both tumours were negative for BRST2. CGH was performed on both the breast and the bladder tumour. The breast tumour showed many aberrations (n = 25), in contrast to the bladder carcinoma, in which only nine chromosomal changes were found. In addition, both tumours showed specific changes—for example, the breast tumour showed amplification at 1q31–32, and in the bladder carcinoma amplifications were present at both telomeres of chromosome 3 (a very rare amplification). This virtually excluded the possibility that both tumours shared a common origin.
Patient 4
At necropsy of a 91 year old woman, multiple tumour localisations were found in the supraclavicular lymph nodes, pancreas, and right breast. The patient had a history of an adenocarcinoma of the left breast for which she underwent a mastectomy 40 years before she died. The tumour in the pancreas was histologically different from the other tumours (fig 5), but immunohistochemistry was inconclusive. All tumour samples were positive for CAM5.2, CK7, AE1/3, and carcinoembryonic antigen (CEA) and negative for BRST2.
Figure 5.
Histology of patient 4. (A) The breast lesion showed adenocarcinoma with predominant tubule formation and moderate cytonuclear atypia, whereas (B) the pancreatic tumour and (C) lymph node metastases both showed poorly differentiated adenocarcinoma, with solid small epithelial nests and moderate to strong nuclear atypia.
CGH analyses of primary breast cancers and metachronous metastases after several decades have been described.23 In this patient, we performed CGH on the breast tumour, the pancreatic tumour, and one lymph node metastasis. Unfortunately, there was no material available from the breast tumour removed 40 years before death. The breast tumour showed 15 chromosomal aberrations, the pancreatic tumour showed nine chromosomal aberrations, and the lymph node metastasis 21. The breast and the pancreatic tumours had seven (of 15 and nine, respectively) events in common. The breast cancer and the lymph node metastasis shared nine (of 15 and 21, respectively) events, and the pancreatic cancer and the lymph node metastasis shared eight (of nine and 21, respectively) events. These CGH results indicated that all the lesions had developed from the same tumour. Based on the idea that the tumour with the least changes is the primary, this would be the pancreatic tumour, although theoretically it is possible that all the tumours found at necropsy had originated from the breast cancer diagnosed 40 years before death. However, clinically, such a long time interval before relapse is less likely.
Patient 5
A 59 year old woman presented with a tumour in the right ovary with clear cell histology. Seven years before she had undergone a nephrectomy because of a renal carcinoma of the left side. Both the combination of a primary renal cell carcinoma and an ovarian clear cell carcinoma, or the alternative, a renal cell carcinoma with a metastasis in the ovary, are rare. Furthermore, the pattern of tumour spread (along the para-aortal lymph nodes with no intra-abdominal metastases) is remarkable for an ovarian adenocarcinoma. Some differences were seen on histology and immunohistochemistry (the renal cell carcinoma was negative for CK7, CEA, CA125, and vimentin, whereas the ovarian tumour was positive for all these markers). CGH showed 11 chromosomal aberrations in the renal cell carcinoma and 25 in the ovarian tumour. Out of the total number of 30 different chromosomal aberrations, the tumours had only five in common. In this case, the combination of genotyping by CGH and immunophenotyping yielded sufficient information to classify the tumours as two independent lesions, and it was therefore concluded that the tumour in the ovary was a second primary.
Patient 6
A 72 year old man who underwent resection of the inferior lobe of the right lung because of a 5.5 cm tumour was found to have an additional 1.1 cm tumour in the same lobe that was quite different histologically. The largest tumour had an epithelial undifferentiated large cell phenotype, whereas the smaller tumour showed a mesenchymal aspect, with polymorphic atypical spindle cells and many mitoses.
The total numbers of chromosomal aberrations detected by CGH were 17 and 19, in the larger and smaller tumours, respectively, of which they had 10 in common. Both lesions showed several chromosomal aberrations that are rather typical for squamous cell carcinomas,12 such as gain of 3q and 11q13, and loss of 13q. Because similar levels of agreement have been seen within tumours, the CGH results were consistent with the two tumours being genetically related.24
Patient 7
A 58 year old woman presented with synchronous endometrioid carcinomas of the ovary and endometrium. Both tumours were diploid as analysed with flow cytometry. CGH was performed on both lesions and revealed only three events in the uterus (8p−, 16q−, and Xp+) and two in the ovarian carcinoma (12q−, 22q−). Because none of these chromosomal aberrations was shared, it was concluded that these two endometrioid tumours were not related, and thus they were considered to be two primary tumours.
DISCUSSION
The traditional role of the pathologist has been the evaluation of human tumour samples in a search for clues to their histogenesis and anticipated clinical behaviour. Over the years, this quest has benefited from the application of specialised techniques, which have become part of the pathologist’s arsenal. In addition to light microscopic evaluation of haematoxylin eosin stained tissue sections, immunohistochemistry has proved to be a particularly valuable adjunct in cancer diagnosis. More recently, molecular pathological techniques have been added to this repertoire of useful techniques.
Irrespective of the tools applied, every diagnosis in clinical pathology, as one of the laboratory disciplines, should be considered as the result of a diagnostic test. For such a test to make an optimal contribution to clinical decision making, ideally it should be validated and implemented in a standardised way.25 This includes defining the algorithm to calculate the classifier, deciding on the optimal cut off point depending on the clinical relevance, and ultimately extensive testing of the performance of the test in terms of positive and negative predictive value. This process is very demanding in terms of labour and availability of clinically and pathologically well documented series of specimens, and consequently it takes many years. When the respective differential diagnoses (which actually means the different diagnostic tests) in every day routine pathology are considered, in the present situation only a subset is large and consistent enough to apply these rules rigorously (for example, testing sentinel nodes for tumour metastases, immunohistochemical panels for discriminating between different types of adenocarcinomas).26,27 A major risk for following this strategy is that by the time the process is completed, an alternative test emerges that is implemented in diagnostic pathology without going through this whole procedure. When we compare this with the situation of putting a new drug on the market, this is a remarkable situation, because the consequences of a substandard test can be as equally severe as the consequences of a substandard drug. If we want to develop clinical pathology as an evidence based branch of medicine, it is obvious that we should strive to extend our repertoire of validated tests.
Nevertheless, in many situations we have to provide answers without having such a validated test available, and we simply use the tools that are available. This can be acceptable as long as both pathologists and clinicians are aware of the limitations of the test used. In the case of obvious test results (for example, the CGH results of two tumours are 100% identical or 100% different) we can provide an answer that has a high degree of certainty, but in all other cases there is a poorly defined level of uncertainty, which should be considered in the ultimate diagnosis and communicated to the clinician who has asked for the test. Against this background, we have explored the possibilities of comparative genomic hybridisation for classifying multiple tumours within one patient as related or independent lesions. It is clear that CGH at this moment does not meet all the criteria described above for a validated diagnostic test—for example, the cut off for deciding whether or not tumours are related is not completely clear. This is further hampered by the fact that tumours may share identical chromosomal changes because they belong to the same histological category. For example, in squamous cell carcinomas loss of 3p, gain of 3q, and amplifications at 11q13 are relatively common, so that finding these changes in two different squamous cell carcinomas provides little information with respect to whether or not the tumours are related. In addition, it can be difficult to draw conclusions when there are relatively small numbers of chromosomal changes. It is more convincing when two tumours share 10 of a total of 20 chromosomal changes, rather than two of four different chromosomal changes. It is essential that we should look for standardisation of test interpretation, preferably based on clear data on the positive and negative predictive value of such tests. However, as with many other tests in pathology, these figures are not readily available, and we should be aware of this fact when interpreting these tests and reporting our conclusions. Nevertheless, when taking these limitations into account, the results of CGH could still contribute towards clinical decision making, as illustrated in the cases presented here.
“It is essential that we should look for standardisation of test interpretation, preferably based on clear data on the positive and negative predictive value of such tests”
When a second tumour presents in the same organ system as the first, the question invariably arises as to whether it is a recurrence or a second primary. The answer to this question may have clinical implications, both with respect to further diagnosis and treatment, and with respect to the patient, for whom a difference can sometimes be made between the presence of metastatic disease versus repeated successful treatment of primary malignancies. Morphological comparison and an extended immunophenotypical profile of different lesions can sometimes resolve the question, but the presence of a clonal mutation or a characteristic pattern of genetic alterations is the most direct way to establish a link between the two lesions, or in contrast, to suggest that the metachronous tumours represent independent events. For this purpose, the pathologist has the choice between testing for the presence of specific mutations (for example, the chance that two independent tumours in one patient share the same p53 mutation is very low),28 using a panel of microsatellite markers,29 or using CGH. Of these, CGH is the only one that tests on a genome wide scale. In our present study, chromosome based CGH has been applied. A new development in this respect is microarray based CGH, one of the “DNA chip” technologies. With this approach, chromosomes are no longer used as a template for hybridisation, but an array of microscopically small spots of genomic DNA is used instead. Every spot represents a unique DNA sequence for which the chromosomal locus is known.30,31 Microarray CGH has a higher resolution and sensitivity, and data analysis is more straightforward than with chromosome based CGH. It is to be expected that this technique will become widely used in routine pathology testing some time in the future.
Take home messages.
Comparative genetic hybridisation (CGH) patterns of gains and losses were useful in supporting the differentiation between metastasis or second primary, and in the identification of the primary tumour location in cases of metastases and two primary tumours in the seven patients analysed
Thus, CGH is a very valuable diagnostic technique in patients with multiple tumours
Microarray CGH has a higher resolution and sensitivity and data analysis is more straightforward than with chromosome based CGH, so that this technique will probably be widely used in routine pathology testing in the future
Abbreviations
CEA, carcinoembryonic antigen
CGH, comparative genomic hybridisation
CK, cytokeratin
REFERENCES
- 1.Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992;258:818–21. [DOI] [PubMed] [Google Scholar]
- 2.Du Manoir S, Speicher MR, Joos S, et al. Detection of complete and partial chromosome gains and losses by comparative genomic hybridization. Hum Genet 1993;90:590–610. [DOI] [PubMed] [Google Scholar]
- 3.Weiss MM, Hermsen MA, Meijer GA, et al. Comparative genomic hybridisation. Mol Pathol 1999;52:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss MM, Hermsen MAJA, Snijders AM, et al. Comparative genomic hybridisation in pathology. In: Crocker J, Murray P. Molecular biology in cellular pathology. Chichester: John Wiley & Sons, 2003:285–305.
- 5.Gebhart E, Liehr T. Patterns of genomic imbalances in human solid tumors. Int J Oncol 2000;16:383–99. [DOI] [PubMed] [Google Scholar]
- 6.Knuutila S, Bjorkqvist AM, Autio K, et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol 1998;152:1107–23. [PMC free article] [PubMed] [Google Scholar]
- 7.Knuutila S, Aalto Y, Autio K, et al. DNA copy number losses in human neoplasms. Am J Pathol 1999;155:683–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grieken van NC, Weiss MM, Meijer GA, et al. Helicobacter pylori-related and -non-related gastric cancers do not differ with respect to chromosomal aberrations. J Pathol 2000;192:301–6. [DOI] [PubMed] [Google Scholar]
- 9.Hermsen M, Guervos MA, Meijer G, et al. New chromosomal regions with high-level amplifications in squamous cell carcinomas of the larynx and pharynx, identified by comparative genomic hybridization. J Pathol 2001;194:177–82. [DOI] [PubMed] [Google Scholar]
- 10.Hermsen MA, Baak JP, Meijer GA, et al. Genetic analysis of 53 lymph node-negative breast carcinomas by CGH and relation to clinical, pathological, morphometric, and DNA cytometric prognostic factors. J Pathol 1998;186:356–62. [DOI] [PubMed] [Google Scholar]
- 11.Hermsen MAJA, Postma C, Baak JPA, et al. Colorectal adenoma to carcinoma progression follows three distinct pathways of chromosomal instability. Gastroenterology 2002;123:1109–19. [DOI] [PubMed] [Google Scholar]
- 12.Weiss MM, Kuipers EJ, Hermsen MAJA, et al. Barrett’s adenocarcinomas resemble adenocarcinomas of the gastric cardia in terms of chromosomal copy number changes, but relate to squamous cell carcinomas of the distal oesophagus with respect to the presence of high-level amplifications. J Pathol 2003;199:157–65. [DOI] [PubMed] [Google Scholar]
- 13.Buerger H, Schmidt H, Beckmann A, et al. Genetic characterisation of invasive breast cancer: a comparison of CGH and PCR based multiplex microsatellite analysis. J Clin Pathol 2001;54:836–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tornoczky T, Kalman E, Jakso P, et al. Solid and papillary epithelial neoplasm arising in heterotopic pancreatic tissue of the mesocolon. J Clin Pathol 2001;54:241–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchio A, Terris B, Meddeb M, et al. Chromosomal abnormalities in liver cell dysplasia detected by comparative genomic hybridisation. Mol Pathol 2001;54:270–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buerger H, Simon R, Schafer KL, et al. Genetic relation of lobular carcinoma in situ, ductal carcinoma in situ, and associated invasive carcinoma of the breast. Mol Pathol 2000;53:118–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szymanska J, Virolainen M, Tarkkanen M, et al. Overrepresentation of 1q21–23 and 12q13–21 in lipoma-like liposarcomas but not in benign lipomas: a comparative genomic hybridization study. Cancer Genet Cytogenet 1997;99:14–18. [DOI] [PubMed] [Google Scholar]
- 18.Simon R, Burger H, Brinkschmidt C, et al. Chromosomal aberrations associated with invasion in papillary superficial bladder cancer. J Pathol 1998;185:345–51. [DOI] [PubMed] [Google Scholar]
- 19.Greulich KM, Utikal J, Peter RU, et al. c-MYC and nodular malignant melanoma. A case report. Cancer 2000;89:97–103. [DOI] [PubMed] [Google Scholar]
- 20.Tienari J, Reima I, Larramendy ML, et al. A cloned human germ cell tumor-derived cell line differentiating in culture. Int J Cancer 1998;77:710–19. [DOI] [PubMed] [Google Scholar]
- 21.Christiaens GC, Vissers J, Poddighe PJ, et al. Comparative genomic hybridization for cytogenetic evaluation of stillbirth. Obstet Gynecol 2000;96:281–6. [DOI] [PubMed] [Google Scholar]
- 22.Daniely M, Aviram-Goldring A, Barkai G, et al. Detection of chromosomal aberration in fetuses arising from recurrent spontaneous abortion by comparative genomic hybridization. Hum Reprod 1998;13:805–9. [DOI] [PubMed] [Google Scholar]
- 23.Kuukasjarvi T, Karhu R, Tanner M, et al. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res 1997;57:1597–604. [PubMed] [Google Scholar]
- 24.Torenbeek R, Hermsen MAJA, Meijer GA, et al. Analysis by comparative genomic hybridization of epithelial and spindle cell components in sarcomatoid carcinoma and carcinosarcoma: histogenetic aspects. J Pathol 1999;189:338–43. [DOI] [PubMed] [Google Scholar]
- 25.Baak JP. The framework of pathology: good laboratory practice by quantitative and molecular methods. J Pathol 2002;198:277–83. [DOI] [PubMed] [Google Scholar]
- 26.Lagendijk JH, Mullink H, van Diest PJ, et al. Immunohistochemical differentiation between primary adenocarcinomas of the ovary and ovarian metastases of colonic and breast origin. Comparison between a statistical and an intuitive approach. J Clin Pathol 1999;52:283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Diest PJ. Histopathological workup of sentinel lymph nodes: how much is enough? J Clin Pathol 1999;52:871–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Sijp Jr, van Meerbeeck JP, Maat AP, et al. Determination of the molecular relationship between multiple tumors within one patient is of clinical importance. J Clin Oncol 2002;20:1105–14. [DOI] [PubMed] [Google Scholar]
- 29.Louhelainen J, Wijkstrom H, Hemminki K. Allelic losses demonstrate monoclonality of multifocal bladder tumors. Int J Cancer 2000;87:522–7. [PubMed] [Google Scholar]
- 30.Snijders AM, Nowak N, Segraves R, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet 2001;29:263–4. [DOI] [PubMed] [Google Scholar]
- 31.Weiss MM, Snijders AM, Kuipers EJ, et al. Determination of amplicon boundaries at 20q13.2 in tissue samples of human gastric adenocarcinomas by high-resolution microarray comparative genomic hybridisation. J Pathol [In press.] [DOI] [PubMed]