Abstract
The Holliday junction is an essential intermediate of homologous recombination. RecA of Bacteria, Rad51 of Eukarya, and RadA of Archaea are structural and functional homologs. These proteins play a pivotal role in the formation of Holliday junctions from two homologous DNA duplexes. RuvC is a specific endonuclease that resolves Holliday junctions in Bacteria. A Holliday junction-resolving activity has been found in both yeast and mammalian cells. To examine whether the paradigm of homologous recombination apply to Archaea, we assayed and found the activity to resolve a synthetic Holliday junction in crude extract of Pyrococcus furiosus cells. The gene, hjc (Holliday junction cleavage), encodes a protein composed of 123 amino acids, whose sequence is not similar to that of any proteins with known function. However, all four archaea, whose total genome sequences have been published, have the homologous genes. The purified Hjc protein cleaved the recombination intermediates formed by RecA in vitro. These results support the notion that the formation and resolution of Holliday junction is the common mechanism of homologous recombination in the three domains of life.
Keywords: DNA recombination, hyperthermophile
Genetic recombination is important in generating genetic diversity and in repairing various types of lethal damage to DNA. The Holliday junction is a critical intermediate structure formed during homologous recombination, in which two homologous duplex DNA molecules are held together by a single-stranded crossover (1). The strand-exchange reaction is mediated mainly by RecA family proteins both in Bacteria and Eukarya. The molecular mechanisms of this early stage of homologous recombination have been studied extensively (reviewed in refs. 2, 3). On the other hand, the mechanism of the late stage of the DNA recombination, in which the Holliday junctions are processed correctly to form the recombinant duplexes, has been analyzed so far mostly in Escherichia coli (reviewed in refs. 4, 5). In E. coli, the RuvA and RuvB complex specifically binds to the Holliday junction and promotes branch migration. RuvC cleaves the Holliday junction when the junction migrates to the favorite sequence of the cleavage. The alternative route of the junction processing by RecG, which has a branch-migration activity similar to that of RuvAB, is not well understood.
The Holliday junction-resolving activity has been identified from a wide variety of organisms from bacteriophage to mammals (reviewed in ref. 6). The biochemical properties of RuvC endonuclease have been well studied. The crystal structure of RuvC was determined at atomic resolution, and the putative model of RuvC complexed with the Holliday junction was also prepared to suggest the cleavage mechanism of DNA (7, 8). The other corresponding enzymes were isolated only from bacteriophage T4 (Endonuclease VII), bacteriophage T7 (Endonuclease I), Lambloid prophage (RusA), and yeast (Endonuclease CCE1). The junction-selective nucleolytic activities have been observed in extracts from mammalian cells (9–12). However, further analyses have failed so far to identify the proteins with the activity and the responsible genes. In yeast, CCE1 is known to be a mitochondrial protein (13). Two more identified activities in yeast called X1 and X3 have not been characterized yet (14, 15). There are homologs of the E. coli ruvA, ruvB, and ruvC genes in the majority of bacterial genomes, although some bacteria lack the homolog of RuvC (16). However, there are no ORFs having similar sequences to bacterial RuvA, RuvB, and RuvC proteins in Eukarya and Archaea in the currently available public databases including the total genomes of Saccharomyces cerevisiae (17) and four archaeal strains, Methanococcus jannaschii (18), Archaeoglobus fulgidus (19), Methanobacterium thermoautotrophicum (20), and Pyrococcus horikoshii (21). Therefore, the question is whether Holliday junctions in the eukaryal and archaeal cells may be processed via novel mechanisms, distinct from that identified in Bacteria, or whether they may be processed by a similar mechanism in Eukarya and Archaea, albeit the sequence of the responsible enzymes may be distinctly different from that of bacterial Ruv proteins.
The RecA/Rad51 structural homologs have been found in Archaea (22), and preliminary characterizations suggest that they are also functional homologs (23, 24). These results show that the archaeal homologs named RadAs are both structurally and functionally similar to the RecA/Rad51 family proteins in the other domains, and they probably play a central role in recombination and repair. The amino acid sequences of archaeal RadAs are more similar to eukaryal Rad51/Dmc1 homologs than to bacterial RecA homologs. From this finding, we expected that the archaeal proteins involved in DNA recombination might be more similar to the eukaryal counterparts and have been trying to identify the proteins involved in the processing of the Holliday junctions in the archaeal cells.
In this study, we identified a Holliday junction-cleaving activity in the hyperthermophilic archaeon, Pyrococcus furiosus, and cloned the gene encoding the protein with the activity. We investigated the general biochemical and physical properties of the purified protein, named Hjc (Holliday junction cleavage).
MATERIALS AND METHODS
DNA Substrates.
Fourteen oligonucleotides (oligos 1–14) were synthesized for making DNA substrates. The sizes of the oligos are: 1–7, 10, and 12, 70 bases; 8 and 9, 59 bases; 11, 60 bases; 13 and 14, 68 bases. Their sequences are available on request. The oligonucleotides were labeled at 5′ termini with [γ-32P] ATP by T4 polynucleotide kinase and were annealed with the complementary oligonucleotides in TAM buffer containing 40 mM Tris⋅acetate (pH 7.8) and 0.5 mM Mg-acetate. Four-way junction with nonhomologous sequence in the junction center (4J) is composed of oligos 1, 2, 3, and 4. Four-way junction with homologous sequence in the core (4Jh) is composed of oligos 2, 5, 6, and 7. Four-way junction with homologous sequences and asymmetrical arm length (4Jhs) is composed of oligos 5, 7, 8, and 9. Three-way junction with nonhomologous arms (3J) and homologous arms (3Jh) are composed of oligos 2, 3, and 13, and oligos 2, 6, and 14, respectively. Duplex DNA (D), Looped-out DNA (L10), and G/A mismatched DNA (G/A) are composed of oligos 2 + 10, 2 + 11, and 2 +12, respectively. Half-cruciform DNAs (Hc1 and Hc2) are composed of oligos 2 + 5 and 2 + 6, respectively. 4Jh, 4Jhs, and 3Jh have a mobile homologous core region.
Endonuclease Assays.
Aliquots (4 μl) of P. furiosus cell extract prepared as described below was incubated with 10 nM of 32P-labeled various substrate DNAs in 36 μl of the standard reaction mixture containing 10 mM Tris⋅HCl, pH8.8, 10 mM MgCl2, 200 mM KCl, and 1 mM DTT at 56°C for 30 min. The reactions were stopped by phenol, and the DNA products were analyzed by PAGE in TAE buffer (40 mM Tris⋅acetate, pH7.8, 1 mM EDTA) or by denaturing PAGE in TBE buffer (90 mM Tris-borate, pH 8.3, 2 mM EDTA).
Purification of the Holliday Junction Cleavage Activity from P. furiosus Cell Extract.
P. furiosus strain Vcl, DSM3638T (25) was cultured as described earlier (26). The cells from 2 liters of culture were suspended in 80 ml of buffer A (50 mM Tris⋅HCl, pH 8.0/0.1 mM EDTA/0.5 mM DTT/10% glycerol) containing 1 mM of PMSF and were disrupted by sonication. After centrifugation of the mixture at 30,000 × g for 20 min at 4°C, proteins (Fraction I) were precipitated with ammonium sulfate (80% saturation) and were dialyzed against buffer A. The dialysate (Fraction II) was applied onto a HiTrap Q column (Amersham Pharmacia) and developed with a 0–1 M NaCl gradient in buffer A. The active fractions eluted at 0.5–0.8 M NaCl were dialyzed against buffer B (10 mM potassium phosphate, pH 6.8/7 mM 2-mercaptoethanol/0.05 mM CaCl2/10% glycerol). The dialysate (Fraction III) was applied onto a hydroxyapatite column (Bio-Rad) and developed with a 0.01–1 M potassium phosphate gradient in buffer B. The active fractions eluted at 0.6–0.8 M potassium phosphate were dialyzed against buffer A (Fraction VI) and applied onto a heparin-Sepharose column (Amersham Pharmacia). This column was developed with a 0–1.5 M NaCl gradient in buffer A, and the activity was eluted at 0.9–1 M NaCl. These fractions were dialyzed against buffer A and stored on ice (Fraction V).
Cloning and Identification of the hjc Gene.
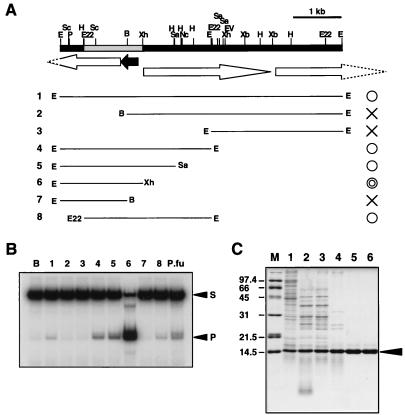
The cosmid-based genomic library of P. furiosus was prepared, and heat-stable protein extracts were obtained from 500 independent clones as described earlier (27), except that heat treatment was performed at 85°C for 10 min. The heat-stable extracts were used for the junction-specific endonuclease assay as described above. Cosmid DNA was prepared from the clone having heat-stable Holliday junction cleavage activity and was partially digested by EcoRI. The DNA fragments were inserted into pUC118, and the resultant plasmids were transformed into E. coli. The heat-stable extracts were prepared from the clones and were used for the junction endonuclease assay. The plasmid named pPFHJ1 containing about a 6-kb fragment shown in Fig. 2 was found to have the activity. To determine the ORF responsible for the activity, deletion mutant series were prepared from pPFHJ1 by digestion with restriction enzymes. Seven fragments shown in Fig. 2 were cut out by indicated enzymes and were inserted into pUC118 (lanes 2 and 7) or pBluescript (lanes 3–6 and 8) vectors. The nucleotide sequences of the DNA fragments were analyzed by a DNA sequencer (ABI Prism 310 genetic analyzer, Applied Biosystems). The sequence of the gene encoding Hjc has been deposited at the DNA DataBank of Japan and has been assigned the accession number AB023635.
Figure 2.
Cloning and expression of the gene responsible for Holliday junction cleavage activity. (A) Restriction map around the gene encoding Holliday junction cleaving enzyme and the results of deletion mutant analysis. B, BamHI; E, EcoRI; Ev, EcoRV; E22, EcoT22I; H, HindIII; Nc, NcoI; Sa, SacI; Sc. ScaI; Sl. SalI; Xb, XbaI; Xh, XhoI. The ORFs found from the nucleotide sequence are indicated by arrows. (B) Result of the Holliday junction cleavage assay by using the heat-treated supernatants derived from the cell extract of E. coli clones that have plasmids carrying each restriction fragment (1–8) indicated in A. The crude extract of P. furiosus was used as a control (P.fu). S and P indicate the substrate and product bands, respectively. (C) SDS/PAGE (15%) analysis of the recombinant Hjc protein from the different stages of the purification. Lanes: 1, supernatant after sonication; 2, polyethylenimine fraction; 3, ammonium sulfate fraction; 4, supernatant after heat treatment; 5, hydroxyapatite fraction; 6, Mono Q fraction. Lane M contains size marker proteins (Bio-Rad). The gel was stained with Coomassie brilliant blue. Arrowhead indicates the Hjc band.
Construction of a Plasmid for Overproduction of Hjc Protein.
The hjc gene was amplified by PCR directly from genomic DNA of P. furiosus. To maintain the accuracy of amplification, Pfu DNA polymerase (Stratagene) was used. Ten nanogram of P. furiosus DNA and 20 pmol of each primer were added to the standard PCR mixture, and 25 cycles with 30 sec each of temperature profile at 94°C, 55°C, and 72°C were performed. The primers are 5′-CGTCGCACGAGCATATGTATAGAAAAG GGGCCC-3′ and 5′-CGCACGAGGATATCTTATCATGATTTCCCCTCCAAC-3′ for forward and reverse, respectively. To adjust the translational initiation codon at the NdeI site of pET21a (Novagen), a NdeI recognition sequence was made in the forward primer. The PCR product was digested with NdeI and inserted into the NdeI-blunt-ended pET21a. The resultant plasmid was designated pPFHJ2.
Purification of Recombinant Hjc Protein.
E. coli BL21(DE3) carrying pPFHJ2 was grown in 2 liters of Luria–Bertani medium containing 100 μg/ml ampicillin to OD600 of 0.7, and the expression of the hjc gene was induced by isopropyl-d-thiogalactoside to a final concentration of 1 mM. After incubation for a further 5 h, the cells were harvested, suspended in 120 ml of buffer A containing 1 mM of PMSF, and disrupted by sonication. The supernatant from centrifugation at 30,000 × g for 20 min was mixed with polyethylenimine at a final concentration of 0.1%. The supernatant was taken by centrifugation, and proteins were precipitated with ammonium sulfate (80% saturation). The precipitate was resuspended in and dialyzed against buffer B and then was treated at 80°C for 30 min. The heat-resistant fraction obtained by centrifugation was applied onto a hydroxyapatite column (Bio-Rad), and it was developed with a 0.01 to 1-M potassium phosphate gradient in buffer B. The fractions eluted at 0.6–0.8 M potassium phosphate contained Hjc protein. These fractions were dialyzed against buffer A and applied onto a MonoQ column (Amersham Pharmacia). The passthrough fraction of the column was directly applied onto a heparin-Sepharose column (Amersham Pharmacia). The chromatography was developed with a 0–1.5 M NaCl gradient in buffer A and the activity eluted at 0.9–1 M NaCl. Peak fractions were dialyzed against buffer A and stored on ice. The concentration of Hjc was determined by using a molar extinction coefficient ɛM of 1.06 × 104 M-1⋅cm−1, which was obtained by a method of Gill and von Hippel (28).
Determination of Hjc Cleavage Sites.
The synthetic Holliday junctions, 4J and 4Jh (10 nM) having uniquely 32P-labeled arm were incubated with or without Fraction V (4 μl) or purified recombinant Hjc (10 nM) at 56°C for 1 hr in standard reaction buffer (40 μl), as described above. The reaction was treated by phenol, and the mixture (8 μl) was mixed with 6 μl of a loading buffer (98% deionized formamide/1 mM EDTA/0.1% xylene cyanol/0.1% bromphenol blue). The products were analyzed by denaturing PAGE and autoradiography.
Resolution of Recombination Intermediates.
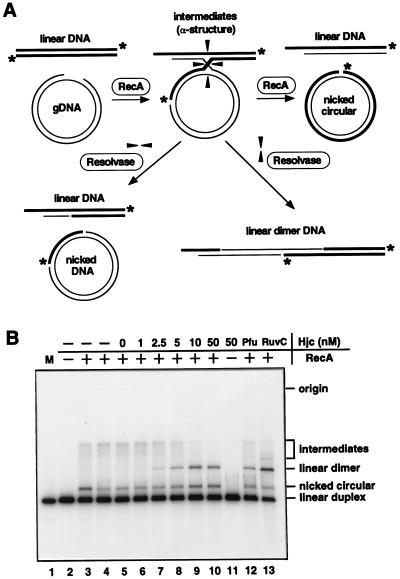
The formation of recombination intermediates by strand-exchange reaction was performed basically as described by Muller et al. (29). For the preparation of the gapped DNA (gDNA), single-stranded circular DNA of plasmid pUC118 and KasI-PstI-digested double-stranded pUC118 were mixed. E. coli RecA protein was prepared as described earlier (30). The plasmid pKM300 for the overproduction of RecA was obtained from T. Horii (Osaka University). RuvC protein was purified by H. Iwasaki (Osaka University) as described earlier (31). RecA-mediated strand-exchange reaction was performed in a buffer containing 20 mM Tris⋅HCl, pH7.5, 15 mM MgCl2, 2 mM DTT, 2 mM ATP, and 100 μg/ml BSA. gDNA (6 μM as nucleotide concentration) was preincubated with RecA protein (2.7 μM) at 37°C for 5 min, and recombination was then initiated by addition of 3′-32P-labeled linear duplex pUC118 (3 μM), which had been cut with PstI. After 20 min of incubation, aliquots were mixed with various concentrations of purified Hjc protein Fraction V from P. furiosus or RuvC (0.17 μM, as a control) and were further incubated at 55°C for 10 min (RuvC, at 37°C for 1 hr). The reaction was stopped by adding SDS (0.5%), EDTA (40 mM), and proteinase K (2 mg/ml) and incubated for an additional 15 min at 37°C and then analyzed by 1.2% agarose gel electrophoresis.
Sedimentation Equilibrium.
Sedimentation equilibrium analysis was performed with a Beckman XL-I Optima Analytical Ultracentrifuge (Beckman Coulter) equipped with absorbance optics. The initial Hjc concentration was 20 μM in buffer A containing 0.1 M of NaCl. The sample was centrifuged at three different speeds, 15,000, 17,000, and 20,000 rpm at 20°C, and the absorbance was monitored at 280 nm. The oligomerization state was determined by fitting the data to a single species, by using Origin Sedimentation Equilibrium Single Data Set Analysis (Beckman Coulter). The partial specific volume used for this analysis was 0.746 ml/g, calculated from the weighted average of the amino acid content by using the method of Cohn and Edsall (32), and the density of the solvent was calculated to be 1.026 g/ml.
Immunodepletion Analysis.
Rabbit polyclonal antibody was raised against homogenous Hjc protein. The polyclonal antisera: anti-Pfu-PCNA, a P. furiosus PCNA protein (I. K. Cann, S. Ishino, I. Hayashi, K.K., H. Toh, K.M. & Y.I., unpublished data) and anti-PI-PfuI, a P. furiosus intein protein (K.K., N. Fujuta, K. Ichiyanagi, H.S., K.M., and Y.I., unpublished data) were used as controls. All subsequent steps were carried out at room temperature as described earlier (33). The immunodepleted extracts were analyzed by endonuclease assay. The precipitants were analyzed by Western blotting by using an enhanced chemiluminescence system (Amersham Pharmacia) according to the method of the supplier.
Computer Analysis of the Amino Acid Sequences.
Search for the homologous sequences in the databases with blast (34) was carried out at a website (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-newblast?Jform=0). clustal w (http://www.clustalw.genome.ad.jp/) was used for the amino acid sequence comparison of the Hjc and other archaeal homologs and for a multiple sequence alignment of them.
RESULTS
Identification and Partial Purification of Junction-Resolving Activity from P. furiosus.
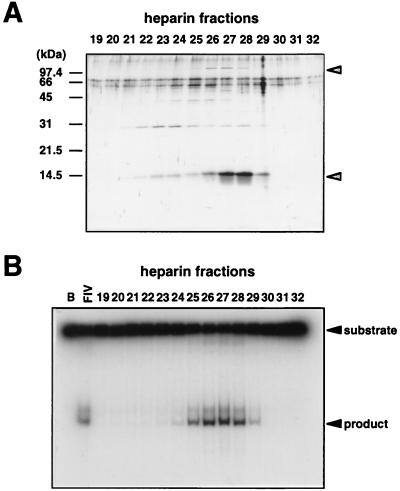
We identified an activity that cleaves a synthetic Holliday junction in the cell extract of P. furiosus after trying various assay conditions. Inclusion of 200 mM KCl in the reaction, which increased the cleavage activity about 4-fold, was critical for the detection of the activity in the crude extracts. The partially purified activity corresponded to a protein band of 14 kDa by the third column chromatography (heparin-Sepharose) as shown in Fig. 1. To examine the substrate specificity of this enzyme activity, we prepared several kinds of DNA substrate, including four-way junctions with or without a central homology core, three-way junctions, half-cruciform, looped-out DNA, and duplex DNA containing one base mismatch. The peak fraction of the activity (Fraction V) cleaved only four-way junctions regardless of the presence of homologous sequences at the junction center (data not shown). No normal duplex DNA with the same sequences of the Holliday junction substrate and the single-stranded DNAs themselves used in this assay was cleaved (data not shown), which suggested that the cleavage activity is not sequence specific.
Figure 1.
Partial purification of Holliday junction cleavage activity from P. furiosus. (A) SDS/PAGE (15%) of the fractions separated by heparin-Sepharose column. All bands in the gel were visualized by silver staining. Protein bands indicated by arrows were eluted in correspondence with the activity. (B) Holliday junction cleavage activity was found in the fractions from the heparin-Sepharose column chromatography. Aliquots of the column fractions (fractions 19–32), together with buffer A (B) and peak fraction from hydroxyapatite chromatography, Fraction IV (IV), were incubated with 32P-labeled 4Jh. The products were analyzed by 12% PAGE followed by autoradiography.
Cloning and Expression of the Gene for the Junction Resolvase.
We screened for the Holliday junction cleaving activity from the heat-stable protein libraries of P. furiosus as described in Materials and Methods. Among 496 independent heat extracts of E. coli transformants, we isolated five clones producing a protein that cleaves specifically the synthetic Holliday junction. All of the five clones contained the same region of P. furiosus genomic DNA in the cosmid vector. As shown in Fig. 2, subcloning, sequencing, and deletion analysis of one of the inserted DNA in the cosmid showed that the shaded region in the map is important and that the ORF shown by the closed arrow is responsible for the activity. The size of the deduced protein from the ORF (13,766 Da) was matched to that of the band detected in the SDS/PAGE of partially purified fraction shown in Fig. 1. Therefore, the gene for the ORF was amplified from the genomic DNA of P. furiosus by PCR and inserted into the expression vector, pET21a, and the nucleotide sequence of the insert DNA was confirmed. The recombinant E. coli BL21(DE3) carrying the resultant plasmid pPFHJ2 was cultivated, and the target protein was successfully overproduced by IPTG induction. Therefore, the protein was purified to homogeneity by the three sequential chromatographies (Fig. 2C). During the purification, we found that the protein has strong affinity for DNA, and polyethylenimine treatment and hydroxyapatite column chromatography were very effective for the separation of the protein from DNA. From 2 liters of E. coli culture, about 5 mg of protein was purified. The highly purified protein was used for the junction cleavage assay, and it was confirmed to have an activity to cleave the junction DNA specifically as described below. From these results, we named it Hjc (Holliday junction cleaving protein).
Immunological Analyses.
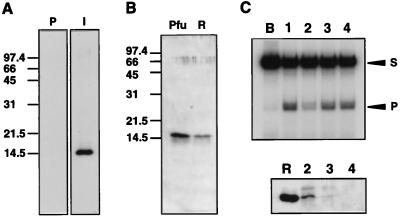
By using highly purified Hjc protein from recombinant E. coli, polyclonal antibody was prepared (Fig. 3A). Western blotting analysis showed that a protein that can specifically react with the antibody was present in the Fraction V of P. furiosus cells (Fig. 3B). The junction-cleaving activity in the cell extract was depleted by treatment with anti-Hjc antibody but not with other antibodies (Fig. 3C). Moreover, a protein corresponding to the size of Hjc was precipitated with anti-Hjc antibody (Fig. 3C). These results strongly support that the activity originally found in the cell extract of P. furiosus was derived from the cloned gene product. We could detect the very faint band of Hjc protein in the crude cell extract of P. furiosus by Western blotting analysis by using the anti-Hjc described above (data not shown). This fact suggests that the amount of Hjc in the cells is not large but its specific activity is high. Fig. 3C shows that Hjc protein was coprecipitated with Pfu-PCNA by anti-Pfu-PCNA (lane 3). This result was reproducible; however, it is not known whether the interaction of the two proteins has some biological meaning.
Figure 3.
Immunological identification of Hjc in P. furiosus cells. (A) The sonication extract of recombinant cells was separated by 15% SDS/PAGE, transferred onto PVDF membranes and treated with immune (I) or preimmune (P) sera raised against Hjc. (B) Fraction V from P. furiosus cells (Pfu) and 17 ng of recombinant Hjc protein (R) were analyzed by Western blotting with anti-Hjc serum. (C) Immunodepletion analysis. (Top) Endonuclease assay of each depleted extract. (Bottom) Each precipitant was analyzed by Western blotting with anti-Hjc serum. Lanes: B, buffer A; 1, crude extract before antiserum treatment; 2, anti-Hjc serum; 3, anti-Pfu-PCNA serum; 4, anti-PI-PfuI serum. Lane R was loaded with 15 ng of recombinant Hjc.
Biochemical Characterization of Hjc Protein.
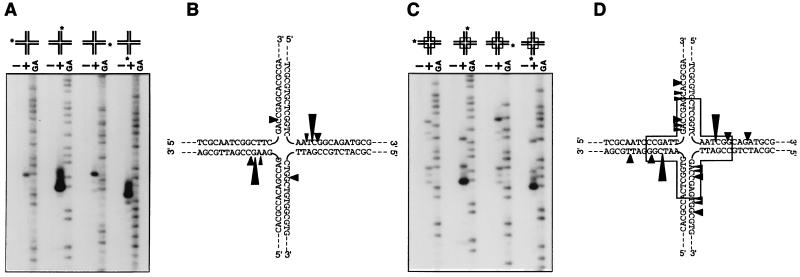
Substrate specificity was analyzed by using the highly purified Hjc protein (Fig. 2C) and the various forms of DNA in the standard assay condition determined above. Hjc cleaved efficiently the four-way junctions. However, it also cleaved three-way junctions (both mobile and immobile) and looped-out DNA substrates, although very inefficiently (data not shown). The exact cleavage sites of the cleavable substrates were determined. As shown in Fig. 4, strands 2 and 4 were mainly cleaved at one specific site in both immobile and mobile four-way junctions, even though other sites, especially in the mobile junction, were cleaved with much less efficiency. These results were exactly the same as those from the partially purified fraction (Fraction V) from P. furiosus cells (data not shown). We observed a pair of dominant cleavage sites located symmetrically across the junction in both mobile and immobile junction, as is the case with other junction resolvases. Minor cleavage sites of the latter were also located symmetrically across the junction. Hjc can cleave mobile and immobile junctions with similar efficiencies. This property is the same as that of a mammalian activity (35) but is in contrast with E. coli RuvC, which requires at least 2 bp of the homology core at the junction center (36).
Figure 4.
Determination of the cleavage sites produced by the Hjc. An immobile four-way junction (A and B) and a mobile junction (C and D), each labeled uniquely at the 5′-32P-end in indicated (∗) strand, were used as substrates. Substrate DNAs were incubated with (+) or without (−) Hjc protein. The GA sequence ladders of each labeled oligonucleotide generated by the Maxam–Gilbert method were loaded alongside to provide markers. The cleavage sites are represented by arrowheads showing cutting efficiencies by their sizes. The box part of the junctions in D indicates the mobile homologous region.
We tested whether the nicked strand in the resolved duplex DNA is rejoined by ligation by using the junction 4Jhs as basically designed, as described earlier (35). Most of the major cleavage products (37 bases) were converted to a longer strand (70 bases) by treatment with T4 DNA ligase (data not shown). This result showed that the cleavage occurred at the symmetrically related sites of the two strands to leave 5′-phosphate and 3′-hydroxyl termini like other junction resolvases.
Resolution by Hjc of Recombination Intermediates Made by RecA Protein.
It is critically important to demonstrate that Hjc can cleave Holliday recombination intermediates made by RecA to show that Hjc is an archaeal functional homolog of E. coli RuvC. Recombination intermediate was formed by using RecA-mediated strand-exchange reaction between gapped circular pUC118 and homologous linear duplex DNA labeled at 3′-termini with 32P. The intermediates of the strand-exchange reaction contained Holliday junctions as shown in Fig. 5A, which were resolved by RuvC to give rise to a linear dimer recombinant product (Fig. 5B, lane 13) as reported previously (37). By using this in vitro recombination system, the resolvase activity of Hjc was tested. As shown in Fig. 5B, the intermediates disappeared and the linear dimer products came up in the concentration-dependent manner of Hjc. The Fraction V from P. furiosus also had the activity to resolve the intermediate (Fig. 5B, lane 12). The nicked circular products made from the cleavage of the other direction also increased in the concentration-dependent manner of Hjc. These results show that Hjc is a functional homolog of RuvC.
Figure 5.
Cleavage of the RecA-mediated recombination intermediate by Hjc. (A) Scheme of the formation of the Holliday junctions by RecA and the resolution of the junctions by resolvase. Asterisks show the 3′-32P-labeled ends. (B) Recombination intermediates between gapped DNA and homologous 32P-labeled linear duplex DNA were formed by RecA and then were treated by indicated concentration of Hjc (lanes 5–11) or Fraction V from P. furiosus (lane 12) at 55°C for 10 min or by 0.17 μM of RuvC (lane 13) at 37°C for 1 hr. The RecA-mediated products that are from incubation of the substrates with RecA for 30 min at 37°C (lane 3) and the intermediates before Hjc treatment (lane 4) were loaded as controls. 32P-labeled linear duplex DNA (3 ng) was loaded as a marker (lane 1). Recombination products were analyzed by 1.2% agarose gel electrophoresis.
Hjc Protein Forms Dimer in Solution.
To determine the native molecular weight of Hjc, we measured sedimentation equilibrium profiles of Hjc protein with a Beckman analytical ultracentrifuge (Beckman Coulter). The apparent molecular weights of Hjc in the buffer A containing 0.1 M NaCl were 25,000, 26,815, and 27,495 Da, respectively, obtained from the three experiments at different speeds. These values are about twice the monomer size Hjc with deduced weight of 13,766 Da. This result indicates that Hjc protein exists as dimer in the solution and probably binds to and cleaves the Holliday junction in the same homodimeric form as other junction resolvases.
Hjc Homologs in Archaea.
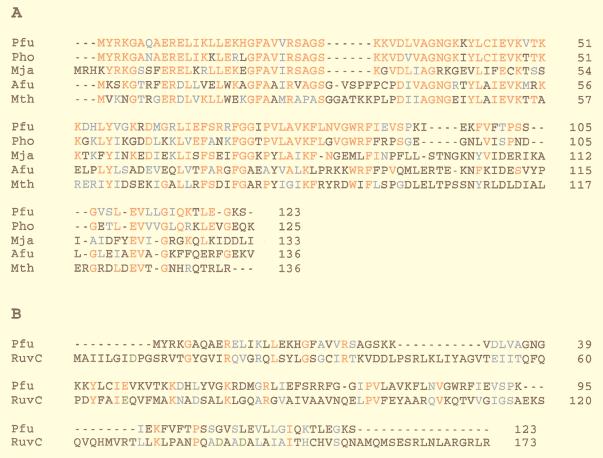
The deduced amino acid sequence of Hjc is not similar to that of any known protein from Eukarya and Bacteria in the public databases. However, ORFs having highly similar sequences were found in the genomes of four archaeal organisms, M. jannaschii, M. thermoautotrophicum, A. fulgidus, and P. horikoshii, all of which genomes have been completely sequenced. The sequence identities among these archaeal strains are 30% on average. The multiple sequence alignments of these proteins are shown in Fig. 6A. To show how different the sequence of Hjc is from that of RuvC, the two sequences were aligned (Fig. 6B). In the Hjc protein family, the N-terminal half-regions are more conserved, compared with the C-terminal half. In contrast, bacterial RuvC proteins are more conserved in the C-terminal region (16). There were several acidic and basic amino acid residues that are completely conserved in the proteins from five archaeal strains. Some of these residues may constitute a catalytic center of the nucleolytic reaction.
Figure 6.
Comparison of Hjc amino acid sequence with homologs found in four archaeal strains (A) and E. coli RuvC (B). Pfu, P. furiosus; Pho, P. horikoshii; Mja, M. jannaschii; Afu, A. fulgidus; Mth, M. thermoautotrophicum. Identical and similar amino acid residues are indicated by red and blue letters, respectively. The four residues indicated by green letters in RuvC are known to constitute the catalytic center (42, 8).
DISCUSSION
This is the first report, to our knowledge, describing a Holliday junction-resolving enzyme in Archaea. The protein presented in this study, which is named Hjc, introduces symmetrically related nicks into two DNA strands of like polarity as observed with RuvC and other known resolvases. Therefore, Hjc can be grouped in the category of Holliday junction resolvases as proposed earlier (35), although it does not share the sequence similarity of any enzymes among them.
Present studies suggest that mechanisms of homologous DNA recombination are fundamentally conserved among three biological domains, Bacteria, Eukarya, and Archaea. Functional and structural homologs of RecA proteins of Bacteria are identified in Eukarya and Archaea, and all of them are shown to be involved in homology search and strand-exchange reaction leading to formation of recombination intermediates (3, 24). Hjc protein efficiently cleaves the recombination intermediate formed by RecA as efficiently as bacterial RuvC Holliday junction resolvase in a similar manner (Fig. 5). These results strongly support that formation and resolution of recombination intermediates are carried out in Archaea by mechanisms similar to those in Bacteria and probably in Eukarya. Formation of Holliday junctions as recombination intermediates have been demonstrated in S. cerevisiae in vivo (38) and activities similar to RuvC have been identified in mammalian cells (35), although the responsible gene for the activity has not been isolated.
We could not find any ORFs having homologous sequence to that of Hjc in the currently available databases. Instead, we found that the sequence of Hjc is clearly conserved in the genomes of euryarchaeotes. This fact shows that the protein must be important for the life of organisms in at least this subdomain of Archaea. The DNA of the hyperthermophilic archaea is known to be extremely resistant to breakage in vivo by radiolysis and thermolysis (39, 40). DiRuggiero et al. showed that the chromosomal DNA in P. furiosus cells was repaired at 95°C after dramatic fragmentation by ionizing radiation (41). These results suggest the presence of very active mechanisms for double-strand break DNA repair in hyperthermophilic Archaea, and Hjc homologs may be some of the crucial enzymes in the process. It would now be very interesting to know whether the protein is also conserved in Crenarchaeota, the other subdomain of Archaea, because most of the organisms found to date in this subdomain are hyperthermophilic. The current sequencing projects of three crenarchaeal strains, Sulfolobus solfataricus, Pyrobaculum aerophilum, and Aeropyrum pernix, will give the answer to this question very soon.
It is necessary to analyze the phenotypes of the hjc mutants to know whether Hjc is actually involved in the homologous recombination and the recombinational repair in vivo. However, no genetic engineering techniques are applicable to P. furiosus. Searching for the homologs in the methanogenic and halophilic Archaea, whose host-vector systems are available, is interesting for functional analyses, such as making disruptants of the hjc gene. It is also very important to find proteins that can promote the branch migration of the Holliday junctions in P. furiosus. Hjc proteins may help as a probe to isolate the proteins related to the branch migration process. The discovery of a Holliday junction resolvase in Archaea sheds light on the understanding of the molecular mechanism in the late stage of DNA recombination in Archaea.
Acknowledgments
We thank Drs. H. Iwasaki and I. Cann for valuable discussions and critical reading of the manuscript. We also thank S. Ishino for preparing RecA protein. We are grateful to Drs. Y. Shimura, D. Söll, and C. Woese for continuous encouragement.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB023635).
References
- 1.Holliday R. Genet Res. 1964;5:282–304. [Google Scholar]
- 2.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinohara A, Ogawa T. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 4.Shinagawa H, Iwasaki H. Trends Biochem Sci. 1996;21:107–111. [PubMed] [Google Scholar]
- 5.West S C. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 6.White M F, Giraud-Panis M-J E, Pohler J R G, Lilley D M J. J Mol Biol. 1997;269:647–664. doi: 10.1006/jmbi.1997.1097. [DOI] [PubMed] [Google Scholar]
- 7.Ariyoshi M, Vassylyev D G, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 8.Morikawa K. In: Nucleic Acids and Molecular Biology. Eckstein F, Lilley D M J, editors. Vol. 12. Berlin: Springer; 1998. pp. 275–299. [Google Scholar]
- 9.Waldman A S, Liskay R M. Nucleic Acids Res. 1988;16:10249–10266. doi: 10.1093/nar/16.21.10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeyaseelan R, Shanmugam G. Biochem Biophys Res Commun. 1988;156:1054–1060. doi: 10.1016/s0006-291x(88)80951-3. [DOI] [PubMed] [Google Scholar]
- 11.Elborough K M, West S C. EMBO J. 1990;9:2931–2936. doi: 10.1002/j.1460-2075.1990.tb07484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solaro P, Greger B, Kemper B. Eur J Biochem. 1995;230:926–933. doi: 10.1111/j.1432-1033.1995.tb20638.x. [DOI] [PubMed] [Google Scholar]
- 13.West S C, Korner A. Proc Natl Acad Sci USA. 1985;82:6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensch F, Kosak H, Seeman N C, Kemper B. EMBO J. 1989;8:4325–4334. doi: 10.1002/j.1460-2075.1989.tb08619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleff S, Kemper B, Sternglanz R. EMBO J. 1992;11:699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hishida T, Iwasaki H, Ishioka K, Shinagawa H. Gene. 1996;182:63–70. doi: 10.1016/s0378-1119(96)00474-x. [DOI] [PubMed] [Google Scholar]
- 17.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel D J, Jacq C, Johnston M, et al. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 18.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 19.Klenk H-P, Clayton R A, Tomb J, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 20.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H-M, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 22.Sandler S J, Satin L H, Samra H S, Clark A J. Nucleic Acids Res. 1996;24:2125–2132. doi: 10.1093/nar/24.11.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods W G, Dyall-Smith M L. Mol Microbiol. 1997;23:791–797. doi: 10.1046/j.1365-2958.1997.2651626.x. [DOI] [PubMed] [Google Scholar]
- 24.Seitz E M, Brockman J P, Sandler S J, Clark A J, Kowalczykowski S C. Genes Dev. 1998;12:1248–1253. doi: 10.1101/gad.12.9.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiala G, Stetter K O. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- 26.Uemori T, Ishino Y, Toh H, Asada K, Kato I. Nucleic Acids Res. 1993;21:259–265. doi: 10.1093/nar/21.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uemori T, Sato Y, Kato I, Doi H, Ishino Y. Genes Cells. 1997;2:499–512. doi: 10.1046/j.1365-2443.1997.1380336.x. [DOI] [PubMed] [Google Scholar]
- 28.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 29.Müller B C, Jones C, Kemper B, West S C. Cell. 1990;60:329–336. doi: 10.1016/0092-8674(90)90747-3. [DOI] [PubMed] [Google Scholar]
- 30.Morimatsu K, Horii T, Takahashi M. Eur J Biochem. 1995;228:779–785. doi: 10.1111/j.1432-1033.1995.tb20323.x. [DOI] [PubMed] [Google Scholar]
- 31.Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohn E J, Edsall J T. Proteins. New York: Reinhold; 1943. pp. 370–381. [Google Scholar]
- 33.Cann I K O, Komori K, Toh H, Kanai S, Ishino Y. Proc Natl Acad Sci USA. 1998;95:14250–14255. doi: 10.1073/pnas.95.24.14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Hyde H, Davies A A, Benson F E, West S C. J Biol Chem. 1994;269:5202–5209. [PubMed] [Google Scholar]
- 36.Shida T, Iwasaki H, Saito A, Kyogoku Y, Shinagawa H. J Biol Chem. 1995;271:26105–26109. doi: 10.1074/jbc.271.42.26105. [DOI] [PubMed] [Google Scholar]
- 37.Dunderdale H J, Benson F E, Paesons C A, Sharples G J, Lloyd R G, West S C. Nature (London) 1991;354:506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- 38.Schwacha A, Kleckner N. Nucleic Acids Res. 1995;24:2125–2132. [Google Scholar]
- 39.Kopylov V M, Bonch-Osmolovskaya E A, Svetlichnyi V A, Miroshnichenko M L, Skobkin V S. Mikrobiologiya. 1993;62:90–95. [Google Scholar]
- 40.Peak M J, Robb F T, Peak J G. J Bacteriol. 1995;177:6316–6318. doi: 10.1128/jb.177.21.6316-6318.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiRuggiero J, Santangelo N, Nackerdien Z, Ravel J, Robb F T. J Bacteriol. 1997;179:4643–4645. doi: 10.1128/jb.179.14.4643-4645.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito A, Iwasaki H, Ariyoshi M, Morikawa K, Shinagawa H. Proc Natl Acad Sci USA. 1995;92:7470–7474. doi: 10.1073/pnas.92.16.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]