Abstract
Aims: To investigate the physical status of human papillomavirus 16 (HPV-16) in low grade squamous intraepithelial lesions (LSILs) as a means of determining the percentage of viral integration.
Methods: Ninety two LSIL/HPV positive Thin Prep™ samples were initially tested for the E6 gene by the polymerase chain reaction (PCR) to identify the HPV-16 virus. To avoid false positive results, the specificity of the bands obtained from PCR was confirmed by Southern blot hybridisation with internal oligonucleotide probes. Next, a PCR screen for the E2 gene was performed to identify those samples in which the virus was integrated. Viral integration was detected in just over half of them.
Results: Twenty of the 92 samples were HPV-16 positive, as shown by PCR for the E6 gene. Southern blot analysis confirmed that 13 of these samples were positive for the viral E6 gene. Thus, viral integration was detected in just over a half of the samples positive for HPV-16.
Conclusions: These data show that HPV-16 integration occurs in a subset of LSILs. The measurement of HPV-16 integration would be a helpful complementary tool for cytological evaluation in primary cervical screening to identify those patients at risk of developing high grade squamous intraepithelial lesions and cervical cancer.
Keywords: human papillomavirus 16, viral integration, low grade squamous intraepithelial lesion
Epidemiological and biochemical data support the division of human papilloma viruses (HPVs) into two groups: high risk (for example, HPV-16, HPV-18, HPV-5, and HPV-8) and low risk (for example, HPV-1, HPV-6, and HPV-11) types,1 and there is considerable evidence showing that cervical dysplasia is induced by persistent infection with high risk HPVs. So far, more than 80 HPV types have been identified and whereas low risk types have been found mostly in benign lesions and low grade squamous intraepithelial lesions (LSILs), HPV-16 and HPV-18 have been categorised as high risk HPV types on the basis of their greater than 50% prevalence in high grade squamous intraepithelial lesions (HSILs) and their 80–90% prevalence in cervical cancers.2–6 HPV-16 is the most well studied HPV type, and serves as an important model for studying viral carcinogenesis. Therefore, there is increased interest in using the detection of HPV DNA as an adjunct to classic cytological evaluation. In fact, the Papanicolaou test is not perfect and false negative rates of 5–50% have been reported for LSILs.7,8 For this reason, in the past few years, the use of liquid cytology has made it possible to combine morphological evaluation with the molecular analysis of the cervical cells.
“There is increased interest in using the detection of human papillomavirus DNA as an adjunct to classic cytological evaluation”
During infection, a subset of several oncoproteins is expressed under a complex regulatory network of cellular and viral transcription factors. Malignant transformation is brought about by the products of the viral E6 and E7 oncogenes, which act by inactivating the tumour suppressor protein p53 and the RB family proteins pRb, p107, and pRb2/p130, respectively.9–16 In HPV-16, the transcription of both the E6 and E7 genes is under the control of the upstream regulatory region and depends on a single promoter, called p97. The full length E2 gene product, a transcriptional activator protein,17,16 represses the transcription of the E6 and E7 genes by binding to the p97 promoter region. It has also been shown that the reintroduction of the E2 protein into HPV-16 transformed cervical carcinoma cells upregulates p97 promoter activity and the cells die via apoptosis.14,18
We could consider integration as a mutation, with consequences for both the cellular and viral genome. In previous studies on chromosomal locations of the HPV-16 genome in cell lines and carcinomas, integration was found mostly in chromosomes 1, 2, 8, 9, 3, 12, 13, and 20.19–22 Selected integrations have been described, by other authors, in common fragile sites,23,24 or in interspersed repetitive sequences of DNA.25 Nevertheless, it has not been possible to demonstrate a specific and preferred integration site in the human genome. From the viral perspective, the integration produces, first of all, a small deletion of DNA, rarely being more than three kilobases. The E2 open reading frame (ORF) has been identified as the preferential site of integration because it has been found to be disrupted or deleted more frequently than other sites.26–30 Therefore, the disruption of E2 dependent negative feedback controlling E6 and E7 transcription is considered a selective event in tumour development and progression.22,28,31,32 Furthermore, several observations have suggested that other E2 functions are necessary to mediate cellular growth arrest. In a recent study,18 the reintroduction of the E2 protein into an HPV-16 transformed cervical carcinoma cell line resulted in a decrease in growth rate and cell death via apoptosis, by increasing the concentration of free E2F. The same effect was previously described in serum starved cells.18,33 These and other data suggest the crucial role of inactivation of the E2 gene by integration and explain why E2 damage is associated with poor prognosis and significantly shorter disease free survival for the patient.34–37
Although some authors have shown the coexistence of episomal and integrated forms in cervical cancer,38–40 according to most data in the literature, viral DNA is usually integrated into the cellular genome in lines derived from cervical carcinomas, in addition to HSILs and invasive cervical carcinomas.6,22,40 Therefore, the phenomenon of integration, leading to progression of dysplasia into carcinoma, is considered to be an important mechanism for tumour progression in the cervix.
In inflammatory states and LSILs, the virus is usually detected as an episomal form.6,21,41 Nevertheless, we focused on LSIL samples because in these early lesions the frequency of integration is still a matter of conjecture.
Several methods have been used to detect integration, such as Southern blot analysis, two dimensional gel electrophoresis, amplification by the polymerase chain reaction (PCR) of the E2/E1 region of the virus, and the ANCHOR PCR, although some of them are relatively insensitive and the results are not always clear. Furthermore, most of these methods require large amounts of DNA and are still too complicated and time consuming for use in daily clinical practice.
We initially tested the samples for the E6 gene by PCR to identify the HPV-16 type virus. The specificity of the bands, obtained from the first PCR, was confirmed by Southern blot hybridisation with internal oligonucleotide probes. Then, a PCR screen for the E2 gene was performed to identify those samples in which the virus was integrated.
MATERIALS AND METHODS
Clinical samples
Residual material from ThinPrep™ samples of 92 LSIL/HPV positive cases was obtained from Thomas Jefferson University Hospital, USA. The diagnosis of LSIL and the presence of HPV were determined after cytological evaluation only, and not confirmed by biopsy because most LSILs are not referred to colposcopy for biopsy. The study population was chosen among young women between 20 and 35 years old with no previous abnormal cytological test. The samples were selected consecutively.
Cytological diagnosis
The samples were prepared for liquid based cytology with the ThinPrep technique (Cytyc Corporation, Marlborough, Massachusetts, USA) and 4 ml of the sample was used to process the slides for cytomorphological evaluation. Samples were classified according to the Bethesda system for reporting cervical/vaginal cytological diagnoses.42 The cytotechnicians and the pathologists involved in our study were not informed about the results of the HPV testing because this was a retrospective study.
HPV-16 testing
The material remaining from the morphological evaluation was used for HPV molecular analysis.
DNA extraction
The cells were centrifuged at 60 000 ×g for 30 minutes, resuspended, and digested in 200 μg/ml proteinase K (Boehringer Mannheim, Mannheim, Germany) for one hour at 37°C. The DNA was extracted with phenol/chloroform, precipitated with ethanol, then dissolved in water and used as a template in PCR reactions.
PCR amplification
Ninety two samples were tested for the presence of HPV-16 by PCR amplification of the E6 gene. The primers used were: 5′-AAGGGCGTAACCGAAATCGGT-3′ and 5′-CATATACCTCACG TCGCAG-3′.43 To detect integration, the DNA from all the 13 samples that had tested positive for the E6 gene at PCR and Southern blot analysis was tested by amplifying the E2 ORF of HPV-16, using specific primers 5′-CTTGGGCACCGAAG AAACAC-3′ and 5′-TTGGTCACGTTGCCATTCAC-3′. The reaction volume was 50 μl and PCR was carried out in the presence of 2.5mM MgCl2 and 0.5 μM of each primer. All the other reagents were used according to the suggestions of the manufacturer. After five minutes at 95°C to denature the DNA, 35 cycles were performed. Each cycle consisted of: denaturation at 95°C for one minute, annealing for one minute, and extension at 72°C for one minute. The annealing steps were performed at temperatures of 58°C for the E2 gene and 60°C for the E6 gene. Samples amplified in the absence of template DNA served as a negative control. SiHa cells were used as a positive control.
Southern blot hybridisation
Southern blot hybridisation was performed on all of the 92 samples to confirm the PCR data for the E6 gene. Aliquots (20 μl) of the E6 gene PCR product were electrophoresed on a 2% agarose gel and transferred overnight in 0.5N NaOH to a positively charged nitrocellulose membrane (Hybond N+; Amersham, Arlington, Illinois, USA). Filter immobilised DNA was prehybridised for two hours at 52°C in a solution containing 5× sodium saline phosphate/EDTA (0.15M NaCl, 0.01M sodium phosphate, 0.001M EDTA, pH 7.7), 5× Denhardt’s solution, 0.5% sodium dodecyl sulfate (SDS), and 100 μg/ml of fresh denatured sheared salmon sperm DNA, and hybridised for 12 hours with a 32P end labelled oligonucleotide, corresponding to the region from nucleotide 136 to nucleotide 161 of the HPV genome sequence, using 1 × 106 counts/minute/ml. The oligonucleotide was labelled with T4 polynucleotide kinase (Promega Corporation, Madison, Wisconsin, USA), according to the manufacturer’s instructions. After hybridisation, the membrane was washed twice in 2× saline sodium citrate (SSC), 0.1% SDS at room temperature for 10 minutes, washed in 0.5× SSC, 0.1% SDS three times at 52°C for 15 minutes, and exposed to x ray.
RESULTS
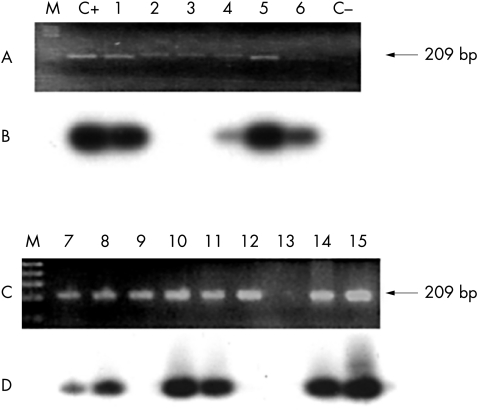
Ninety two samples cytomorphologically diagnosed as LSIL/HPV positive were analysed. The E6 gene was detected in 20 samples by PCR, which is in accordance with the data concerning the proportion of high risk HPVs found in LSIL.44 Southern blot hybridisation was performed on the same samples and 13 of 92 were confirmed as positive (fig 1).
Figure 1.
Identification of human papillomavirus 16 (HPV-16) in low grade squamous intraepithelial lesions. Ninety two samples were tested for the presence of HPV-16 by polymerase chain reaction (PCR) amplification of the E6 gene. (A,C) Some of the samples positive for E6 are shown (lanes 1–15). C+ and C− correspond to the positive and negative controls, respectively. Arrows on the right show the expected molecular weight of the E6 gene fragment (209 bp). (B,D) The E6 gene PCR product (20 μl) was electrophoresed on a 2% agarose gel and hybridised using a single strand oligonucleotide corresponding to the region of the HPV genome spanning nucleotides 136 to 161.
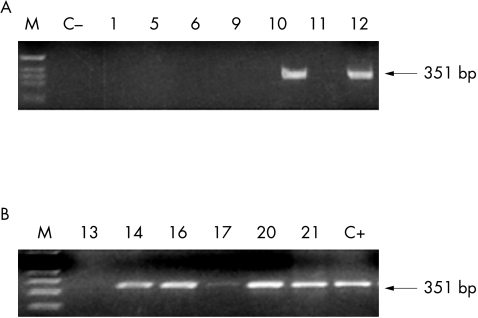
We analysed these 13 samples for integration and in seven we could not detect the E2 gene PCR product, suggesting that the virus was integrated into the cellular genome of these HPV-16 positive cases. Figure 2 shows the E2 PCR products of some of the 13 samples that were positive by both PCR and Southern blot for the E6 gene, and hence thought to be HPV-16 positive.
Figure 2.
Study of viral integration. For the detection of integration, the E2 open reading frame of HPV-16 was amplified. Examples of viral genomic integration (lanes 1, 5, 6, 9, 11, and 13) and of viral episomal forms (lanes 10, 12, 14, 16, 17, 20, and 21) are shown. Arrows on the right show the expected molecular weight of the E2 gene fragment (351 bp).
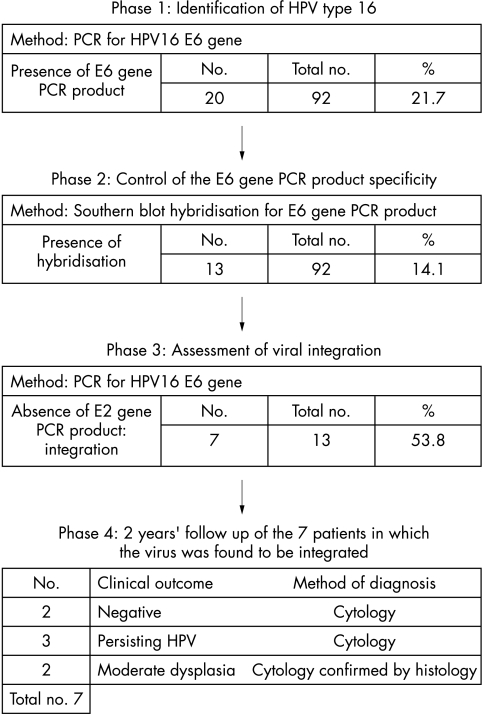
We followed the patients in whom the virus was found to be integrated for two years by cytological evaluation. Two of them had negative cytology, three had persistent HPV infection, and the remaining two showed moderate dysplasia confirmed by histology. The data are summarised in fig 3.
Figure 3.
Diagnostic method of identification of human papillomavirus 16 (HPV-16) and the detection of viral integration. The figure shows, in a temporal sequence, the steps followed in HPV-16 identification and in the detection of viral integration, ending with the follow up. Results of each phase of the study (numbered from 1 to 4) are shown below. In phase 1, HPV-16 was detected by polymerase chain reaction (PCR) for the E6 gene and 20 of the 92 cases were positive. In phase 2, Southern blot hybridisation was performed on the E6 gene PCR product as a control. Thirteen of the 92 cases were confirmed to be HPV-16 positive. Phase 3 was the study of viral integration among the HPV-16 positive cases. The absence of E2 gene PCR product implies viral integration and this was found in seven of the 13. In the last phase, the clinical outcome is summarised (after two years of follow up) for those patients in whom the virus had been integrated. In five of these seven patients, HPV infection or moderate dysplasia persisted during the two following years.
The use of Southern blot analysis has several advantages: it allows confirmation of the type of HPV identified, which was type 16 in those samples in which we found more than one PCR product; it decreases the risk of false negatives because it can detect small amounts of DNA, which are often undetectable by ethidium bromide staining (fig 1; lane 6); and moreover, this method reduces the risk of obtaining false positives because it makes it possible to discriminate small differences in size between two PCR products (fig 1; lane 9). For example, in the primary screening, 20 of the 92 samples were found to be HPV-16 positive, whereas Southern blot analysis allowed us to determine that only 13 of these samples were really positive.
DISCUSSION
It is now accepted that the integration of high risk HPVs into the host cell genome is one of the major contributing factors to genital malignant transformation. To provide a better understanding of this complex phenomenon, it would be interesting to establish temporal relations in HPV induced carcinogenesis. Most authors agree with the hypothesis that the integration of the HPV genome takes place very early in the development of cancer. The question is: how early? Considering viral integration as the key point in cervical carcinogenesis, we wanted to investigate the physical status of HPV-16 in LSILs.
We focused on type 16 HPV because its behaviour seems to differ from that of other high risk papilloma viruses. As shown in a recent study,40 cervical carcinoma reveals differences in the integration profile depending on the virus type. For HPV-18, HPV-31, and HPV-35, the viral genome is always present in the integrated form, whereas for HPV-16 the episomal and integrated forms coexist even in cancer,22,38–41,45 and the concordance between integration and carcinogenesis is less evident. Moreover, we were interested in LSIL because the correlation between cytological and histological findings is often poor in the diagnosis of LSIL. The literature reports a rate of cytological overdiagnosis of 15% and underdiagnosis of between 28% and 62%.46–49 In addition, there is currently no consensus as to the appropriate management of women with this kind of lesion; opinions include immediate colposcopy and directed biopsy, as with cytological HSILs, follow up with repeat cytology every four to six months and colposcopy (indicated only if an abnormality persists), or triage using DNA testing for cancer associated HPV types.50,51 Therefore, we chose to investigate the frequency of integration of a high risk HPV (HPV-16) in LSIL.
We found that HPV-16 was integrated in more than half of the cases that were HPV-16 positive. These results contrast with most of the studies in the literature41,45 that have looked at inflammatory states and LSIL, in which the virus is usually detected in its episomal form, as a 8 kb circular molecule. We agree that viral integration is a very early event, as already postulated by most authors, but our results show that it occurs earlier than the onset of morphological changes, which could indicate a high grade lesion. In attempts to explain our data, we suggest that molecular events precede morphological features leading to malignancy, and that integration does not always temporally coincide with a high grade lesion. It is also possible that viral integration is not necessarily always followed by immediate viral oncoprotein expression. Nevertheless, our data concerning the persistence of HPV or LSIL after two years of follow up in five of seven patients carrying the integrated virus lead us to suppose that integration represents a point of “no return” in the natural history of the lesion.
Take home messages.
Viral integration was detected in just over half of the samples positive for human papillomavirus type 16 (HPV-16)
The measurement of HPV-16 integration would be a helpful complementary tool for cytological evaluation in primary cervical screening to identify those patients at risk of developing high grade squamous intraepithelial lesions and cervical cancer
“We agree that viral integration is a very early event, as already postulated by most authors, but our results show that it occurs earlier than the onset of morphological changes, which could indicate a high grade lesion”
Several techniques have been described to detect the physical status of HPV-16, such as Southern blot analysis, two dimensional gel electrophoresis, amplification by PCR of the E2/E1 region of the virus, and ANCHOR PCR. However, the interpretation of the results remains difficult. In the procedure we describe, we initially tested the samples for the E6 gene by PCR to identify the HPV-16 type virus. The specificity of the bands obtained from the first PCR was confirmed by Southern blot hybridisation with internal oligonucleotide probes on all of the samples. Then a PCR screen for the E2 gene was performed to identify those samples that were HPV-16 positive in which the virus is integrated. This method has several advantages: it allows the detection of integration using a small amount of DNA (0.3–1.0 mg); the interpretation of the results is easier than with other techniques because it is based on the presence or absence of PCR products; furthermore, the results obtained by the PCR technique are confirmed by Southern blot hybridisation analysis.
These days, cytomorphological evaluation alone is not thought to be sufficient for grading cervical dysplasia, and these results need to be supported by molecular HPV testing. In this regard, liquid based cytology has the double advantage of having a sensitivity significantly higher than that of conventional cytology (87.8% v 68.1%; p < 0.05)52–55; moreover, it allows us to combine the morphological examination with molecular HPV testing.
Very recently, most authors have focused their attention on the issue of whether HPV testing might be of value in primary screening or in the assessment of defined patient groups with borderline changes and mild dyskaryosis.46,56 For example, the combination of liquid based cytology and HPV testing enhances sensitivity to ensure that all patients with dyskaryotic lesions are identified. Nevertheless, sensitivity and specificity are generally inversely related to one another, and the higher sensitivity could lead to an increase in the detection of HPV DNA even in patients without detectable disease. Therefore, if HPV testing only provides us with data about the presence or absence of the virus, it is not as useful as a test that also provides information about the physical status (integration) of the virus in the host cell.
Our strategy appears to be a helpful complementary tool for cytological evaluation because it can reduce unnecessary colposcopy guided biopsies in women with a cytological diagnosis of LSIL and might help identify those patients who are at risk for developing HSIL and cervical cancer. Moreover, we suggest the use of this technique in clinical practice.
Acknowledgments
We thank M Basso for her editorial assistance during the preparation of this manuscript. This work was supported in part by grants from the NIH, the Sbarro Health Research Organisation (AG), a grant from the Instituto Pasteur Fondazione Cenci Bolognetti (GG), and a Dottorato di Ricerca in Biologica Diagnostica Quantitativa from the University of Siena (LB).
Abbreviations
HPV, human papillomavirus
HSIL, high grade squamous intraepithelial lesion
LSIL, low grade squamous intraepithelial lesion
ORF, open reading frame
PCR, polymerase chain reaction
SCC, saline sodium citrate
SDS, sodium dodecyl sulfate
REFERENCES
- 1.Zwerschke W, Jansen-Dürr P. Cell transformation by the E7 oncoprotein of human papillomavirus type 16: interactions with nuclear and cytoplasmic target proteins. Adv Cancer Res 2000;78:1–29. [DOI] [PubMed] [Google Scholar]
- 2.Arends MJ, Wyllie AH, Bird CC. Papillomaviruses and human cancer. Hum Pathol 1990;21:686–98. [DOI] [PubMed] [Google Scholar]
- 3.Arends MJ, Donaldson YK, Duvall E, et al. HPV in full thickness cervical biopsies: high prevalence in CIN 2 and CIN 3 detected by a sensitive PCR method. J Pathol 1991;165:301–9. [DOI] [PubMed] [Google Scholar]
- 4.Arends MJ, Donaldson YK, Duvall E, et al. HPV 18 associates with a more advanced cervical neoplasia than HPV 16. Hum Pathol 1993;24:432–7. [DOI] [PubMed] [Google Scholar]
- 5.Stanley M. Genital papillomaviruses, polymerase chain reaction and cervical cancer. Genitourin Med 1990;66:415–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donaldson YK, Arends MJ, Duvall E, et al. A PCR approach to discriminate between integrated and episomal HPV DNA in small clinical specimens. Mol Cell Probes 1993;7:285–92. [DOI] [PubMed] [Google Scholar]
- 7.Schneider A, Zahm DM, Kirchmayr R, et al. Screening for cervical intraepithelial neoplasia grade 2/3: validity of cytologic study, cervicography, and human papillomavirus detection. Am J Obstet Gynecol 1996;174:1534–41. [DOI] [PubMed] [Google Scholar]
- 8.Cuzick J, Terry G, Ho L, et al. Type-specific human papillomavirus DNA in abnormal smears as a predictor of high-grade cervical intraepithelial neoplasia. Br J Cancer 1994;69:167–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyson N, Howley PM, Münger K, et al. The human papillomavirus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 1989;243:934–7. [DOI] [PubMed] [Google Scholar]
- 10.Hawley-Nelson P, Vousden KH, Hubbert NL, et al. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J 1989;8:3905–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Münger K, Phelps WC, Bubb V, et al. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol 1989;63:4417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe S, Kanda T, Yoshiike K. Human papillomavirus transformation of primary human embryonic fibroblast requires expression of open reading frames E6 and E7. J Virol 1989;63:965–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 1990;248:76–9. [DOI] [PubMed] [Google Scholar]
- 14.Lewis H, Webster K, Sanchez-Perez A-M, et al. Cellular transcription factors regulate human papillomavirus type 16 gene expression by binding to a subset of the DNA sequences recognized by the viral E2 protein. J Gen Virol 1999;80:2087–96. [DOI] [PubMed] [Google Scholar]
- 15.Smith-McCune K, Kalman D, Robbins C, et al. Intranuclear localization of human papillomavirus 16 E7 during transformation amd preferential binding of E7 to the Rb family member p130. Proc Natl Acad Sci U S A 1999;96:6999–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishiji T. Molecular mechanism of carcinogenesis by human papillomavirus-16. J Dermatol 2000;27:73–86. [DOI] [PubMed] [Google Scholar]
- 17.Ushikai M, Lace MJ, Yamakawa Y, et al. Transactivation by the full-length E2 proteins of human papillomavirus type 16 and bovine papillomavirus type 1 in vitro and in vivo: cooperation with activation domains of cellular transcription factors. J Virol 1994;68:6655–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Perez A-M, Soriano S, Clarke AR, et al. Disruption of the human papillomavirus type 16 E2 gene protects cervical carcinoma cells from E2F-induced apoptosis. J Gen Virol 1997;78:3009–18. [DOI] [PubMed] [Google Scholar]
- 19.Choo KB, Chen CM, Han CP, et al. Molecular analysis of cellular loci disrupted by papillomavirus 16 integration in cervical cancer: frequent viral integration in topologically destabilized and transcriptionally active chromosomal regions. J Med Virol 1996;49:15–22. [DOI] [PubMed] [Google Scholar]
- 20.Couturier J, Sastre-Garau X, Schneider-Maunoury S, et al. Integration of papillomavirus DNA near myc genes in genital carcinomas and its consequences for proto-oncogene expression. J Virol 1991;65:4534–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durst M, Croce CM, Gissamann L, et al. Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc Natl Acad Sci U S A 1987;84:1070–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalantari M, Blennow E, Hagmar B, et al. Physical state of HPV16 and chromosomal mapping of the integrated form in cervical carcinomas. Diagn Mol Pathol 2001;10:46–54. [DOI] [PubMed] [Google Scholar]
- 23.Thorland EC, Myers SL, Persing DH, et al. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res 2000;60:5916–21. [PubMed] [Google Scholar]
- 24.Bauer-Hofmann R, Borghouts C, Auvinen E, et al. Genomic cloning and characterization of the nonoccupied allele corresponding to the integration site of human papillomavirus type 16 DNA in the cervical cancer cell line SiHa. Virology 1996;217:33–41. [DOI] [PubMed] [Google Scholar]
- 25.Carmody MW, Jones M, Tarraza H, et al. Use of the polymerase chain reaction to specifically amplify integrated HPV-16 DNA by virtue of its linkage to interspersed repetitive DNA. Mol Cell Probes 1996;10:107–16. [DOI] [PubMed] [Google Scholar]
- 26.Baker CC, Phelps WC, Lindgren V, et al. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol 1987;61:962–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choo KB, Pan CC, Han S. Integration of human papillomavirus type 16 into cellular DNA of cervical carcinoma: preferential deletion of the E2 gene and invariable retention of the long control region and the E6/E7 open reading frames. Virology 1987;161:259–61. [DOI] [PubMed] [Google Scholar]
- 28.zur Hausen H. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology 1991;184:9–13. [DOI] [PubMed] [Google Scholar]
- 29.IARC Working Group. IARC monographs on the evaluation of carcinogenic risks to humans. Human papillomaviruses. Lyon: IARC Scientific Publication 64, 1995. [PMC free article] [PubMed]
- 30.Badaracco G, Venuti A, Sedati A, et al. HPV16 and HPV18 in genital tumors: significantly different levels of viral integration and correlation to tumor invasiveness. J Med Virol 2002;67:574–82. [DOI] [PubMed] [Google Scholar]
- 31.Romanczuk H, Howley PM. Disruption of either the E1 or E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc Natl Acad Sci U S A 1992;9:3159–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells SL, Francis DA, Karpova AY, et al. Papillomavirus E2 induces senescence in HPV-positive cells via pRB- and p21 (CIP)-dependent pathways. EMBO J 2000;19:5762–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci U S A 1994;91:3602–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unger ER, Vernon SD, Thoms WW, et al. Human papillomavirus and disease-free survival in FIGO stage Ib cervical cancer. Int J Cancer 1995;74:1184–90. [DOI] [PubMed] [Google Scholar]
- 35.Vernon SD, Unger ER, Miller DL, et al. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer. Int J Cancer 1997;74:50–6. [DOI] [PubMed] [Google Scholar]
- 36.Kalantari M, Karlsen F, Kristensen G, et al. Disruption of the E1 and E2 reading frames of HPV 16 in cervical carcinoma is associated with poor prognosis. Int J Gynecol Pathol 1998;17:146–53. [DOI] [PubMed] [Google Scholar]
- 37.Lazo PA. The molecular genetics of cervical carcinoma. Br J Cancer 1999;80:2008–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kristiansen E, Jenkins A, Holm R. Coexistence of episomal and integrated HPV16 DNA in squamous cell carcinoma of the cervix. J Clin Pathol 1994;47:253–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park JS, Hwang ES, Park SN, et al. Physical status and expression of HPV genes in cervical cancers. Gynecol Oncol 1997;65:121–9. [DOI] [PubMed] [Google Scholar]
- 40.Pirami L, Giache V, Becciolini A. Analysis of HPV16, 18, 31, and 35 DNA in pre-invasive and invasive lesions of the uterine cervix. J Clin Pathol 1997;50:600–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dürst M, Kleinheinz A, Hotz M, et al. The physical state of human papillomavirus type 16 DNA in benign and malignant genital tumours. J Gen Virol 1985;66:1515–22. [DOI] [PubMed] [Google Scholar]
- 42.National Cancer Institute Workshop. The revised Bethesda system for reporting cervical/vaginal cytologic diagnoses. Acta Cytol 1993;37:115–24. [PubMed] [Google Scholar]
- 43.Yoshinouchi M, Hongo A, Nakamura K, et al. Analysis by multiplex PCR of the physical status of human papillomavirus type 16 DNA in cervical cancers. J Clin Microbiol 1999;37:5929–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh YL, Shin KJ, Han J, et al. Significance of high-risk human papillomavirus detection by polymerase chain reaction in primary cervical cancer screening. Cytopathology 2001;12:75–83. [DOI] [PubMed] [Google Scholar]
- 45.Daniel B, Mukherjee G, Seshadri L, et al. Changes in the physical state and expression of human papillomavirus type 16 in the progression of cervical intraepithelial neoplasia lesions analysed by PCR. J Gen Virol 1995;76(Pt 10):2589–93. [DOI] [PubMed] [Google Scholar]
- 46.Bolger S, Lewis BV. A prospective study of colposcopy in women with mild dyscariosis or koilocytosis. Br J Obstet Gynaecol 1988;95:1117. [DOI] [PubMed] [Google Scholar]
- 47.Nyirjesy I. Atypical or suspicious cervical smears: an aggressive diagnostic approach. J Am Med Assoc 1972;222:691. [PubMed] [Google Scholar]
- 48.Davis RM, Cooke JK, Kirk RF. Cervical conization: an experience with 400 patients. Obstet Gynecol 1972;40:23. [PubMed] [Google Scholar]
- 49.Lee SSN, Collins RJ, Pun TC, et al. Conservative treatment of low grade squamous intraepithelial lesions (LSIL) of the cervix. Int J Gynaecol Obstet 1998;60:35–40. [DOI] [PubMed] [Google Scholar]
- 50.Kurman RJ, Henson DE, Herbst AL, et al. Interim guidelines for management of abnormal cervical cytology. The 1992 National Cancer Institute workshop. JAMA 1994;271:1866–9. [PubMed] [Google Scholar]
- 51.Solomon D, Schiffman M, Tarone R. Comparison of three management strategies for patients with atypical squamous cells of undetermined significance: baseline results from a randomized trial. J Natl Cancer Inst 2001;93:293–9. [DOI] [PubMed] [Google Scholar]
- 52.Clavel C, Masure M, Bory J-P, et al. Human papillomavirus testing in primary screening for the detection of high-grade cervical lesions: a study of 7932 women. Br J Cancer 2001;89:1616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Papillo JL, Zarka MA, St-John TL, et al. Evaluation of the ThinPrep Pap test in clinical practice. A seven-month 16314-case experience in Northern Vermont. Acta Cytol 1998;42:203–8. [DOI] [PubMed] [Google Scholar]
- 54.Sherman ME, Mendoza M, Lee KR et al. Performance of liquid-based, thin layer cervical cytology: correlation with reference diagnoses and human papillomavirus testing. Mod Pathol 1998;11:837–43. [PubMed] [Google Scholar]
- 55.Weintraub J, Morabia A. Efficacy of a liquid-based thin layer method for cervical cancer screening in a population with low incidence of cervical cancer. Diagn Cytopathol 2000;22:55–9. [DOI] [PubMed] [Google Scholar]
- 56.Herrington CS. Does HPV testing have a role in primary cervical screening? Cytopathology 2001;12:71–4. [DOI] [PubMed] [Google Scholar]