Abstract
Background: Bilaterality in breast cancer is a rare event and together with an early onset of disease points towards inheritance of the disease. However, most cases seem to occur sporadically, either in a synchronous or metachronous manner.
Methods: Thirty two invasive carcinomas and one in situ carcinoma from 16 patients with synchronous, bilateral breast cancer were investigated by means of comparative genomic hybridisation (CGH) and polymerase chain reaction based multiplex microsatellite analysis. The results were analysed conventionally and were also subjected to a biomathematical cluster analysis.
Results: On average, bilateral breast cancer cases showed a low number of genetic alterations, a low frequency of genetic amplifications, and a high rate of chromosomal 16q losses. A distinct, characteristic genetic alteration associated with bilateral breast disease could not be found. Although two tumour pairs appeared to be related using biomathematical processing for microsatellite analysis, this result was reproduced by CGH data processing in one patient only.
Conclusions: Most synchronous, bilateral breast cancer cases seem to represent independent tumours rather than metastatic events. Nevertheless, the possibility of a specific susceptibility remains.
Keywords: breast cancer, bilateral, comparative genomic hybridisation, microsatellites, cluster analysis
Theoretically, from a tumour biological point of view, the coincidental occurrence of two independent malignant tumours within one organ or an organ system, such as the human breast, is a rather improbable event. In clinical practice, 5–10% of patients with breast cancer will suffer from bilateral tumours, predominantly metachronous disease, associated with a higher frequency of multicentricity, whereas only 1% of patients with breast cancer present with synchronous bilateral breast cancer. The roles of a positive family history or histological differentiation in the likelihood of acquiring bilateral breast cancer disease are controversial. Nevertheless, a young age of onset of disease points to an inherited, familial background, and patients with breast cancer who have BRCA 1 and BRCA2 germ line mutations are included in this category.1 However, no differences in the overall and disease free survival rates could be detected in both subgroups in contrast to unilateral disease.2
“The roles of a positive family history or histological differentiation in the likelihood of acquiring bilateral breast cancer disease are controversial”
Results of immunohistochemical and genetic investigations have provided evidence to support the two different hypotheses concerning the evolution of synchronous bilateral breast cancer. Whereas conventional cytogenetic investigations showed the presence of identical balanced chromosomal alterations in bilateral breast cancer, indicating that these tumours result from a metastatic event,3 other investigators provided evidence for the independent pathogenesis of these tumours.4
We aim to provide evidence that most synchronous bilateral breast cancer cases result from two tumours arising independently. Comparative genomic hybridisation (CGH), as a method to gain an overview of all unbalanced chromosomal alterations within a tumour, in combination with polymerase chain reaction (PCR) based multiplex microsatellite analysis and biomathematical cluster analysis, are an ideal combination of techniques to determine the degree of clonal association between synchronous bilateral breast cancer cases.
METHODS AND MATERIALS
Materials
Our study comprised 16 patients with synchronous bilateral breast cancers, defined as when both tumours were diagnosed within a time period of six months. All patients were diagnosed between 1997 and 2001. The average age was 68.5 years (range, 41–89; median, 68). Staging was performed using the criteria of the TNM system, and grading was done according to established protocols.5 Eight invasive carcinomas were graded as grade 1, 17 as grade 2, and seven as grade 3. Fifteen carcinomas were lymph node positive.
CGH analysis
CGH analysis and the evaluation of genetic alterations were performed as described previously.6,7 Only metaphase spreads showing an even, high intensity hybridisation with low granularity were taken into account. Corresponding ratio profiles were evaluated only if the 95% confidence limits did not exceed 0.15. The 50% thresholds (upper threshold, 1.25; lower threshold, 0.75) were applied to define the chromosomal regions of DNA sequence losses or gains. Independent confirmation of chromosomal aberrations has shown that these thresholds are reliable and eliminate the possibility of false positive results. The consistency of these aberrations has been confirmed by previous reverse CGH experiments (tumour DNA labelled with digoxigenin; reference DNA labelled with biotin). Each CGH experiment included a control hybridisation of fluorescein isothiocyanate and rhodamine labelled normal DNA to each other.
DNA was isolated from paraffin wax embedded material. If necessary, at least 25 sections of 10 μm thickness were manually microdissected under microscopic control.
PCR based multiplex microsatellite analysis
All breast lesions were analysed by means of PCR based multiplex microsatellite analysis using a panel of 11 polymorphic markers and using the same DNA as for CGH. Reference DNA was isolated from paraffin wax embedded, tumour free axillary lymph nodes of each patient.
PCR assays of epidermal growth factor receptor (EGFR) (forward primer, 5′-(5-FAM)GTT TGA AGA ATT TGA GCC AAC C-3′; reverse primer, 5′-TTC TTC TGC ACA CTT GGC AC-3′) and p53 (forward primer, 5′-(5-FAM)AAG AAA TTC CCA CTG CCA CTC-3′; reverse primer, 5′-ATC CCC TGA GGG ATA CTA TTC-3′) were performed in 10 μl reactions containing 1× PCR buffer II (Perkin Elmer, Foster City, California, USA), 2mM MgCl2, 50 μM of each GeneAmp™ dNTP (Perkin Elmer), 1 μM of forward and reverse primer, 30 ng template DNA, and 0.5 U Ampli Taq Gold (Perkin Elmer). The other nine markers were grouped in three multiplex PCRs of three markers each with the following variations of the primer concentration: multiplex 1 (D7S522 forward primer, 5′-(5-FAM)GCA GGA CAT GAG ATG ACT GA-3′; D7S522 reverse primer, 5′-GTT ATG CCA CTC CCT CAC AC-3′; D8S258 forward primer, 5′-(5-FAM)AGC TGC CAG GAA TCA ACT GAG AG -3′; D8S258 reverse primer, 5′-GAT GCT CAC ATA AAG GAG GGA GG -3′; D16S400 forward primer, 5′-(5-FAM)GGT TCA CAA TTG GAC AGT AT-3′; D16S400 reverse primer, 5′-GAA CCC TCC ATG CTG ACA TT-3′) and multiplex 2 (NEFL forward primer, 5′-(5-FAM)CCA ATA CCT GCA GTA GTG CC-3′; NEFL reverse primer, 5′-GAG CTG CTT AAC ACA TAG GG-3′; D13S153 forward primer, 5′-(5-FAM)AGG GTT ATG TAT AAC CGA CTC C-3′; D13S153 reverse primer, 5′-GTC TAA GCC CTC GAG TTG TGG-3′; D17S855 forward primer, 5′-(5-FAM)GGA TGG CCT TTT AGA AAG TGG-3′; D17S855 reverse primer, 5′-ACA CAG ACT TGT CCT ACT GCC-3′) worked with 0.3 μM of each downstream and upstream primer, whereas multiplex 3 showed a reliable performance only with 0.2 μM of both D10S541 (forward primer, 5′-(5-FAM)CAC CAC AGA CAT CTC ACA ACC-3′; reverse primer, 5′-CCA GTG AAT AGT TCA GGG ATG G-3′), 0.3 μM of both D16S402 (forward primer, 5′-(5-FAM)GT ACC CAT GTA CCC CCA ATA-3′; reverse primer, 5′-CAA AGC ACC ACA TAG ACT AA-3′), and 0.5 μM of both D16S422 (forward primer, 5′-GAG AGG AAG GTG GAA ATA CA-3′; reverse primer, 5′-GTT TAG CAG AAT GAG AAT AT-3′) primers. The PCR reactions were overlaid with mineral oil and carried out in the presence of one fluorescence labelled primer for each microsatellite marker in a 96 well thermocycler (GeneAmp™ PCR System 9700; PE Applied Biosystems). A denaturation step at 95°C for 10 minutes was followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, primer extension at 72°C for 30 seconds, and one final extension at 72°C for seven minutes.
Samples (1–2 μl) of the amplified PCR products were diluted in 20 μl water (high performance liquid chromatography grade) containing 0.5 μl GENESCAN™ 400 HD [Rox] fluorescent size standard (ABI; Foster City, California, USA). The mix was denatured at 95°C for two minutes and cooled for at least 10 minutes at 4°C.
Separation of the PCR generated alleles was performed by the ABI PRISM™ 3700 DNA analyser (ABI) using the Polymer 3700 POP-6TM and 1× 3700 running buffer + EDTA (ABI). The data were analysed by means of the GeneScan analysis software 3.5. To standardise the analysis, the loss of heterozygosity (LOH) score was calculated according to Canzian et al.8
Statistical tests
Biomathematical analysis of the results was performed by producing an Euclidean distance metric of the result vectors and ordering the results by agglomerative hierarchical clustering (complete linkage). The results were assembled in vectors, representing an ordered view of observations from chromosome 1 arm q to chromosome X (for example, chromosome 1 q +, chromosome 1 q−, chromosome 1 p +, etc) or from processed microsatellites (for example, BB1/2, D7S522, etc). LOH was counted as one event, irrespective of the allele affected.
Distance matrices of these vectors give a measure of the relatedness of feature vectors, consisting of the observable features of one case/patient. Cluster analysis was used to produce similarity groups out of the distance matrices. This approach was used because of the good agglomerative coefficient, which is an indicator of the amount of clustering structure found, and the comparability with other result sets. The methods used are part of the mathematical system SPlus6.
RESULTS
CGH analysis
On average, 4.9 alterations/case (range, 0–13) were found in the invasive breast cancer cases.
Chromosomal gains were most commonly seen on 1q (58%), 8q (30%), 17q (16%), and 20q (14%). Chromosomal regions commonly involved in chromosomal losses were 2q (19%), 6q (33%), 8p (22%), 11q (25%), 16q (52%), and 17p (22%). In the patient with associated ductal carcinoma in situ (DCIS; patient 4602), identical CGH ratio profiles (1q+ and 16q−) were obtained from all three tumours (two tubular invasive carcinomas, ductal carcinoma grade 1; fig 1). All other tumour pairs revealed dissimilar CGH ratio profiles
Figure 1.
Comparative genomic hybridisation (CGH) profile and corresponding microsatellite analysis of chromosome 16q (microsatellite markers: D16S400 and D16S402) from case 4602. All tumours revealed identical chromosomal aberrations (1q+, 16q−) in the CGH ratio profile. Microsatellite analysis revealed that both right and left sided invasive tumours (inv.; right, 4602A2; left, 4602B) showed concordant loss of heterozygosity and loss of the same 16q alleles, whereas the DCIS component showed a loss of the other alleles. Loss of the shorter allele, black arrow; loss of the longer allele, white arrow.
LOH analysis
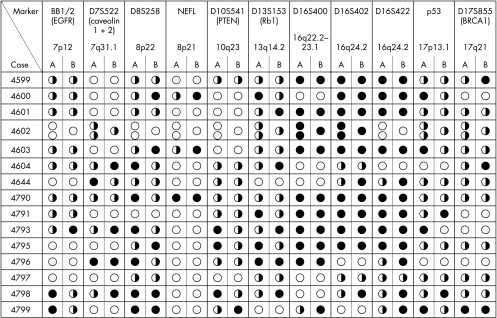
DNAs originating from 16 patients with bilateral breast cancer were analysed. Frequencies of LOH varied from 15% (EGFR) to 95% (D16S400), with a median frequency of 43%. In detail: EGFR, 15% (three cases with LOH and 20 heterozygous); caveolin 1+2, 43% (six cases with LOH and 14 heterozygous); D8S258, 46% (11 cases with LOH and 24 heterozygous); NEFL, 67% (four cases with LOH and six heterozygous); PTEN, 20% (four cases with LOH and 20 heterozygous); Rb1, 35% (nine cases with LOH and 26 heterozygous); D16S400, 95% (19 cases with LOH and 20 heterozygous); D16S402, 77% (20 cases with LOH and 26 heterozygous); D16S422, 77% (20 cases with LOH and 26 heterozygous); p53, 23% (six cases with LOH and 26 heterozygous); and BRCA1, 18% (four cases with LOH and 22 heterozygous) (fig 2). The LOH pattern was identical for all microsatellite markers in one patient only.
Figure 2.
Results of loss of heterozygosity (LOH) analysis of right (A) and left (B) breast cancers for all the microsatellite markers used. Open circles, uninformative; half closed and half open circles, no LOH; closed circles, LOH. If more than one circle is listed, the corresponding nomenclature is A1, A2, etc.
In all 32 invasive breast cancer cases, simultaneous LOH affecting the same genetic locus was seen. The same allele was affected in 23 cases only.
Cluster analysis for LOH and CGH data
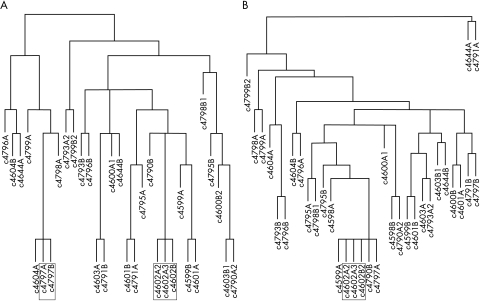
Cluster analysis of both the CGH and microsatellite analysis data revealed tree-like structures. Two tumour pairs were found to be almost identical or related (cases 4602 and 4797) in LOH analysis. Clustering analysis of the CGH data showed close cytogenetic similarity in only one patient (case 4602; fig 3A, B).
Figure 3.
(A) Cluster analysis (Eucledian distance matrix, complete linkage) of loss of heterozygosity data. (B) Cluster analysis of comparative genomic hybridisation data. Closely related tumour pairs are framed.
The two tumours found to be similar by CGH clustering were both staged as lymph node negative. Morphologically, the two tumours from patient 4797 were classified as ductal invasive and lobular invasive carcinomas. In patient 4602, both invasive tumours were classified as tubular invasive carcinomas, whereas the DCIS component was highly differentiated. CGH analysis showed that all three tumours had an identical combination of 16q loss and 1q gain. Microsatellite analysis for 16q markers in this patient revealed loss of the same allele in both invasive tumours, in contrast to the associated DCIS, which showed loss of the other allele.
DISCUSSION
Bilateral breast cancer accounts for 3–4% of all breast cancer cases,2 and might be interpreted as the extreme form of multifocal breast cancer disease. Only a few reports have dealt with genetic findings in sporadic, synchronous breast cancer, concentrating on different aspects and factors in breast carcinogenesis.
The clinical history of our patients provided no evidence to suggest that our series might include cases of familial breast cancer, and the high mean age of our patients supports this assumption. It was also interesting that the average number of genetic alterations seen in each case was lower than that reported in unilateral, sporadic breast cancer.9,10 In addition, the rate of 16q losses as another indicator of tumour grade,11 and the low number of tumours with high level chromosomal gains revealed a high degree of homology with well differentiated ductal invasive tumours, such as well and intermediately differentiated DCIS.12 Nevertheless, because these cytogenetic differences were not significant (data not shown), it is not possible to distinguish between unilateral and bilateral breast cancer cases on a cytogenetic basis alone.
“Comparative genomic hybridisation offers the opportunity to gain an overview of all unbalanced chromosomal alterations within a given tumour, thereby providing a single tumour specific cytogenetic fingerprint”
Distinguishing between bilateral and unilateral breast cancer is also not possible using a “higher resolution” technique, such as microsatellite analysis, because chromosomal regions harbouring the responsible genes were not affected at an increased frequency, as in unilateral breast cancers.10,13 The frequencies of LOH in the respective alleles in our tumour series were comparable to those described in the literature.7,14–18
It is still not clear whether bilateral breast cancers have an independent origin or are the result of a breast to breast metastasis sequence. CGH offers the opportunity to gain an overview of all unbalanced chromosomal alterations within a given tumour, thereby providing a single tumour specific “cytogenetic fingerprint” with a high, but limited, number of parameters investigated. A major drawback of this technique is that balanced translocations within breast cancer cannot be detected, and that bilateral breast cancer cases resulting from a putative breast to breast metastasis sequence based on such translocations will be missed.3 Identical CGH ratio profiles were found in the in situ and invasive tumour parts on both sides in only one patient. Nevertheless, these results are usually interpreted subjectively and biomathematical procedures provide the possibility for a more objective evaluation of the genetic data. Various algorithms to solve such tasks have been described. The above mentioned algorithm has been chosen after a careful review of our results and comparison with the results of similar algorithms. Using hierarchial clustering for the CGH results, the tumours of one patient were clustered as almost identical (fig 2B) and another as possibly related events. In the patient with the highest degree of cytogenetic homology in the left and right sided tumours, only a 1q gain and a 16q loss were seen in the CGH ratio profile, indicating an unbalanced t(1;16) translocation as the underlying mechanism for this combination of chromosomal gains and losses. However, the fact that these three tumours had identical 1q/16q alterations cannot be regarded as proof of a common clonal origin, because the combination of 1q gains and 16q losses, sometimes as sole detectable cytogenetic abnormalities, has been described in up to 30–40% of all sporadic breast cancer cases.19 In addition, the finding of this combination of cytogenetic alterations in bilateral, multifocal lobular carcinoma in situ20 points to a breast tissue specific susceptibility, reflected in a specific chromosomal translocation. The microsatellite analysis data substantiate this hypothesis because, surprisingly, both invasive tumours showed loss of the same 16q alleles, whereas the DCIS component showed a loss of the other alleles (D16S400 and D16S402; fig 1). The clinicopathological findings of T1 and lymph node negative, invasive carcinomas in this patient provide further evidence against a breast to breast metastatic sequence. In contrast, the other tumour (case 4797) clustered as similar by LOH analysis showed a ductal invasive and a pure lobular invasive growth pattern.
In conclusion, in agreement with existing literature using different methods,21,22 and irrespective of whether the results are interpreted by conventional or biomathematical means, synchronous bilateral breast cancer cases represent a spectrum of only partially inherited diseases, with different underlying pathogenetic mechanisms. Most of these cases are independent tumours, and they may result from a breast specific chromosomal instability.
Take home messages.
Most synchronous cases of bilateral breast cancer result from the development of independent tumours rather than metastatic events
However, it is possible that a breast specific susceptibility exists
Table 1.
Overview of all unbalanced chromosomal alterations in bilateral breast cancer determined by comparative genomic hybridisation
| Case | Chromosomal gains | Chromosomal losses |
| 4598A | 1q | 22 |
| 4598B | 8q | 8pter–21; 12q22–ter; 16q21–ter |
| 4599A | 1q | 16q |
| 4599B | 16q | |
| 4600A1 | 16 | 3pter–13; 13q11–31 |
| 4600B2 | 1q; 8q | 6q24–ter; 8pter–21 |
| 4601A | 1q | 2q23–ter; 6q;18q |
| 4601B | 6q11–16; 16q12.1–ter | |
| 4602A2 | 1q21–ter | 16q12.2–ter |
| 4602A3 | 1q21–ter | 16q |
| 4602B | 1q21–ter | 16q12.2–ter |
| 4603A | 11q21–ter; 16q22–ter; 22q11.2–ter | |
| 4603B1 | 8q | 2q22–32; 7q; 16q12.2–ter; 22q12–ter |
| 4604A | 1q21–ter; 8p21–11.1; 14q11.1–22; 17q; 20; 22 | 6q; 11q13–ter |
| 4604B | 3pter–24; 8q; 11p14–11.1 | 2q; 6q14–ter; 17p |
| 4644A | 1q; 3; 5p14–11; 7; 8; 10pter–11.2; 11; 20; 21 | 5q13–ter; 9; 14q; 18 |
| 4644B | 1q21-ter | 2q14.2–24; 4p; 16q12.2–ter; 17p; 22 |
| 4790A2 | 8q | 8pter–22; 16q21–ter |
| 4790B | 1q | 16q; 22p |
| 4791A | 1; 4p; 6p; 11q14-ter; 15q11.2-15; 20 | 2; 3pter–22; 4q21–ter; 6q; 11q14–ter; 13q; 18; 21 |
| 4791B | 1q | 2q11.1–32; 6p23–qter; 8p; 16q |
| 4793A2 | 16q; 17; 22q | |
| 4793B | 20 | 6q; 22qter |
| 4795A | 1q | 6q16–ter; 11q21–ter; 16q |
| 4795B | 1q; 11q12–13; 17q11.1–21 | |
| 4796A | 8q; 17 | 2q22–32; 6q; 13q |
| 4796B | 20 | 6q |
| 4797A | 1q | 16q |
| 4797B | 1q | 3p22–14; 8pter–12; 16q |
| 4798A | 1q; 8q | 8p; 17p; 18 |
| 4798B1 | 1q | 11q14–ter; 16q; 17pter–q12 |
| 4799A | 1q; 5p; 7; 17q21–ter | 11q13–ter; 12p; 17p; 18 |
| 4799B2 | 8p21–qter; 12pter–q21; 13q11–21 | 8pter–22; 10q11.1–23; 11q14–ter; 12q23–ter; 16q |
Acknowledgments
We thank U Neubert, P Meier, and L Grote for technical assistance in CGH analysis. This work was supported by grant Deutsche Krebshilfe 10–1681-Bü-I.
Abbreviations
CGH, comparative genomic hybridisation
DCIS, ductal carcinoma in situ
EGF, epidermal growth factor receptor
LOH, loss of heterozygosity
PCR, polymerase chain reaction
REFERENCES
- 1.Kollias J, Man S, Marafie M, et al. Loss of heterozygosity in bilateral breast cancer. Breast Cancer Res Treat 2000;64:241–51. [DOI] [PubMed] [Google Scholar]
- 2.Mose S, Adamietz IA, Thilmann C, et al. Bilateral breast carcinoma versus unilateral disease. Review of 498 patients. Am J Clin Oncol 1997;20:541–5. [DOI] [PubMed] [Google Scholar]
- 3.Pandis N, Teixeira MR, Gerdes AM, et al. Chromosome abnormalities in bilateral breast carcinomas. Cytogenetic evaluation of the clonal origin of multiple primary tumors. Cancer 1995;76:250–8. [DOI] [PubMed] [Google Scholar]
- 4.Dawson PJ, Maloney T, Gimotty P, et al. Bilateral breast cancer; one disease or two? Breast Cancer Res Treat 1991;19:233–44. [DOI] [PubMed] [Google Scholar]
- 5.Elston CW, Ellis IO. The breast. Edinburgh: Churchill Livingstone, 1998.
- 6.Kallioniemi A, Kallioniemi, OP, Piper J, et al. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci U S A 1994;91:2156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buerger H, Gebhardt F, Schmidt H, et al. Length and loss of heterozygosity of an intron 1 polymorphic sequence of egfr is related to cytogenetic alterations and EGFR-expression. Cancer Res 2000;60:854–7. [PubMed] [Google Scholar]
- 8.Canzian F, Salovaara R, Hemminki A, et al. Semiautomated assessment of loss of heterozygosity and replication error in tumors. Cancer Res 1996;56:3331–7. [PubMed] [Google Scholar]
- 9.Buerger H, Otterbach F, Simon R, et al. Different genetic pathways in the evolution of invasive breast cancer are associated with distinct morphological subtypes. J Pathol 1999;189:521–6. [DOI] [PubMed] [Google Scholar]
- 10.Tirkkonen M, Tanner M, Karhu R, et al. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer 1998;21:177–84. [PubMed] [Google Scholar]
- 11.Buerger H, Mommers E, Littmann R, et al. Correlation of morphologic and cytogenetic parameters of genetic instability with chromosomal alterations in in situ carcinomas of the breast. Am J Clin Pathol 2000;114:854–9. [DOI] [PubMed] [Google Scholar]
- 12.Buerger H, Otterbach F, Simon R, et al. Comparative genomic hybridization of ductal carcinoma in situ of the breast—evidence of multiple genetic pathways. J Pathol 1999;187:396–402. [DOI] [PubMed] [Google Scholar]
- 13.Imyanitov EN, Togo AV, Suspitsin EN, et al. Evidence for microsatellite instability in bilateral breast carcinomas. Cancer Lett 2000;154:9–17. [DOI] [PubMed] [Google Scholar]
- 14.Hampl M, Hampl J, Reiss G, et al. Loss of heterozygosity accumulation in primary breast carcinomas and additionally in corresponding distant metastases is associated with poor clinical outcome. Clin Cancer Res 1999;5:1417–25. [PubMed] [Google Scholar]
- 15.Zenklusen JC, Bieche I, Lidereau R, et al. (C-A)n microsatellite repeat D7S522 is the most commonly deleted region in human primary breast cancer. Proc Natl Acad Sci U S A 1994;91:12155–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia JM, Silva JM, Dominguez G, et al. Allelic loss of the PTEN region (10q23) in breast carcinomas of poor pathophenotype. Breast Cancer Res Treat 1999;57:237–43. [DOI] [PubMed] [Google Scholar]
- 17.Querzoli P, Albonico G, Grazia di Iasio M, et al. Biophenotypes and survival of BRCA1 and TP53 deleted breast cancer in young women. Breast Cancer Res Treat 2001;66:135–42. [DOI] [PubMed] [Google Scholar]
- 18.Hanby AM, Kelsell D, Potts HW, et al. Association between loss of heterozygosity of BRCA1 and BRCA2 and morphological attributes of sporadic breast cancer. Int J Cancer 2000;88:204–8. [PubMed] [Google Scholar]
- 19.Tsuda H, Takarabe T, Fukutomi T, et al. der(16)t(1;16)/der(1;16) in breast cancer detected by fluorescence in situ hybridization is an indicator of better patient prognosis. Genes Chromosomes Cancer 1999;24:72–7. [DOI] [PubMed] [Google Scholar]
- 20.Lu YJ, Osin P, Lakhani SR, et al. Comparative genomic hybridization analysis of lobular carcinoma in situ and atypical lobular hyperplasia and potential roles for gains and losses of genetic material in breast neoplasia. Cancer Res 1998;58:4721–7. [PubMed] [Google Scholar]
- 21.Shibata A, Tsai YC, Press MF, et al. Clonal analysis of bilateral breast cancer. Clin Cancer Res 1996;2:743–8. [PubMed] [Google Scholar]
- 22.Imyanitov EN, Suspitsin EN, Grigoriev M, et al. Concordance of allelic imbalance profiles in synchronous and metachronous bilateral breast carcinomas. Int J Cancer 2002;100:557–64. [DOI] [PubMed] [Google Scholar]