Abstract
Aims: To detect Simian virus 40 (SV40) DNA in mesotheliomas from New Zealand and from England using novel real time FRET probe polymerase chain reaction (PCR) protocols.
Methods: Twenty four mesotheliomas from New Zealand (Central North Island) and 32 mesotheliomas from England (Greater Manchester region) were examined. Two real time FRET probe PCR protocols were optimised and their analytical sensitivity compared using dilutions of SV40 DNA. A conventional SV40 large tumour antigen protocol with detection by probe hybridisation and chemiluminescent Southern blotting was also optimised.
Results: Both real time PCR protocols had the same analytical sensitivity, detecting down to 10−6 pg of SV40 DNA for each reaction, approximately one SV40 copy. All of the 56 mesothelioma samples contained amplifiable β globin DNA, but none contained amplifiable SV40 DNA with the conventional large T antigen PCR–Southern blotting protocol, or the two real time FRET probe PCR protocols. The positive and negative controls gave the expected results. There was no evidence of inhibition.
Conclusions: There is abundant evidence in the literature for the presence of SV40 in mesotheliomas. However, this study found no evidence of SV40 in mesotheliomas from England and New Zealand. The extensive use of SV40 contaminated polio vaccine in New Zealand does not seem to have resulted in SV40 associated mesotheliomas.
Keywords: mesothelioma, Simian virus 40, oncogenesis, polymerase chain reaction, virus
The detection of Simian virus 40 (SV40) DNA in mesotheliomas was first reported in 1994 in the USA.1 Subsequently, numerous studies have also detected SV40 in mesotheliomas and certain other human tumours.2,3 To date, not only has SV40 DNA been repeatedly detected in mesotheliomas, but there have also been related observations, such as the expression of the SV40 large tumour antigen in mesothelioma cells.4 It is now generally accepted that SV40 virus DNA can be found in mesotheliomas, although there are a few recent studies that have produced opposing evidence.2
Our study set out to detect SV40 DNA in mesotheliomas from New Zealand and from England using novel real time FRET probe polymerase chain reaction (PCR) protocols.
METHODS
Our study took place in an International Accreditation New Zealand (IANZ) accredited diagnostic pathology laboratory in a tertiary referral hospital.
Twenty four pleural mesotheliomas from New Zealand (Central North Island) and 32 pleural mesotheliomas from England (Greater Manchester region) were examined. Formalin fixed, paraffin wax embedded tissue was used to prepare haematoxylin and eosin stained sections and sections that were stained immunohistochemically for cytokeratin 7 (CK7), CK5/6, calretinin, desmin, carcinoembryonic antigen, Ber-EP4, B72.3, and CD15. Each of the paraffin wax embedded blocks selected contained abundant tumour, sufficient for numerous serial 5 μm thick sections. A histopathologist experienced in the diagnosis of mesothelioma then reviewed these cases.
One molecular biology technician carried out all of the extractions and amplifications.
DNA extraction
DNA was extracted from 5 μm thick paraffin wax embedded sections using the High Pure PCR template preparation kit (Roche Molecular Biochemicals, Auckland, New Zealand), according to the manufacturer’s protocol for isolation of DNA from formalin fixed, paraffin wax embedded tissue, except that 0.08 mg/ml of poly A carrier RNA was added to the elution buffer and the final elution volume was 50 μl. This modified nucleic acid isolation method involved the lysis of 25 mg of the dewaxed tissue in proteinase K, followed by binding of nucleic acids to the surface of glass fibres in the presence of a chaotrophic salt. The bound nucleic acids were purified from salts, proteins, and other impurities by two washing steps and were eluted in a low salt buffer. This method has been specifically designed for the isolation of viral nucleic acids for clinical diagnostic testing.
PCR
Two real time FRET probe PCR protocols were optimised and their analytical sensitivity compared using dilutions of SV40 DNA (Invitrogen Ltd, Auckland, New Zealand). A conventional SV40 large T cell antigen protocol with detection by probe hybridisation and chemiluminescent Southern blotting5 was also optimised. The control SV40 DNA used was a 5243 bp supercoiled circular DNA isolated from SV40 strain 776.
Real time PCR protocol 1
Protocol 1 was a real time FRET probe PCR protocol using PCR primers SV40S (5′-TTG CTG TGC TTA CTG AGG ATG-3′) and SV40A (5′-CCA ATT ATG TCA CAC CAC AGA-3′), which amplify a 154 bp region within the SV40 large T cell antigen gene. The two FRET probes used for real time detection within this region were: SV40 LCR (5′-LC red-TCA ACC CAC ACA AGT GGA TCT TTC CT -3′-P) and fluorescein labelled SV40 FLU (5′-CAC ATT CTA AAG CAA TCG AAG CAG TAG C-X-3′). A 4 μl aliquot of template DNA was added to a 20 μl reaction mix, which contained 2 μl of 10× LightCycler FastStart DNA master hybridisation probes mix (Roche Molecular Biochemicals), 4mM MgCl2 (total concentration), 0.5 μM of each primer oligonucleotide, and 0.2 μM of each hybridisation probe oligonucleotide. Amplification was performed on a Roche LightCycler (Roche Molecular Biochemicals) using the following protocol: initial denaturation at 95°C for 10 minutes to activate the FastStart Taq DNA polymerase, 50 PCR cycles consisting of heating at 20°C/second to 95°C, with a 10 second hold; cooling at 20°C/second to 60°C, with a 10 second hold; and heating at 20°C/second to 72°C, with a 20 second hold. Fluorescence values of each capillary were measured at 640 nm (channel 2).
Real time PCR protocol 2
Protocol 2 was a real time FRET probe PCR protocol using LC red labelled PCR primer SV40iLC (5′-GTC ACA CCA CAG AAG XTAA–3′) and unlabelled primer SV40 F (5′-GTG CTT ACT GAG GAT GAA–3′), which amplify a 124 bp region within the large T antigen gene of SV40. The fluorescein labelled FRET probe used for real time detection within this region was: SV40-FL (5′-TGG ACT TGA TCT TTG TGA AGG AAC X-3′). A 4 μl aliquot of template DNA was added to a 20 μl reaction mix, which contained 2 μl of 10× LightCycler FastStart DNA master hybridisation probes mix, 3mM MgCl2 (total concentration), 0.5 μM of each primer oligonucleotide, and 0.15 μM of hybridisation probe oligonucleotide. Amplification was performed on a Roche LightCycler using the following protocol: initial denaturation at 95°C for 10 minutes to activate the FastStart Taq DNA polymerase, 50 PCR cycles consisting of heating at 20°C/second to 95°C, with a 10 second hold; then 20°C/second cooling to touchdown annealing from 56°C, with a 0.2°C reduction in annealing temperature each cycle until 50°C, with a 10 second hold at each cycle; and heating at 20°C/second to 72°C, with a 20 second hold. Fluorescence values of each capillary were measured at 640 nm (channel 2).
Conventional large T antigen PCR–Southern blotting protocol
The protocol used was as previously published by Pacini et al,5 using primers originally designed by Bergsagel et al,6 a dioxigenin labelled probe (5′-XGGA AAG TCC TTG GGG TCT TCT ACC–3′), and the CPD-Star™ chemiluminescent detection system (Roche Applied Science, Auckland, New Zealand). The primers amplify a 104 bp region within the large T antigen gene of SV40.
Inhibition control
To determine the presence of Taq DNA polymerase inhibitors, all mesothelioma samples were tested using the Roche LightCycler Control kit (Roche Applied Science). This kit amplifies a 110 bp fragment of the human β globin gene. The target sequence was detected in real time using the double stranded DNA binding dye SYBR green I.
RESULTS
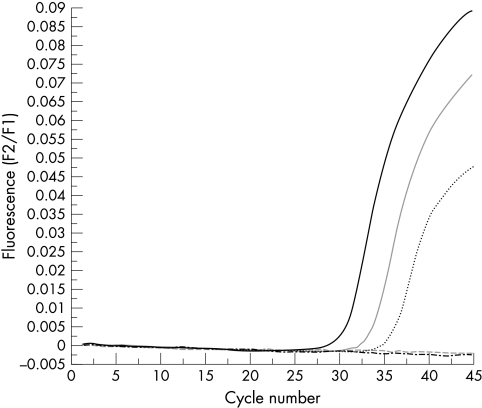
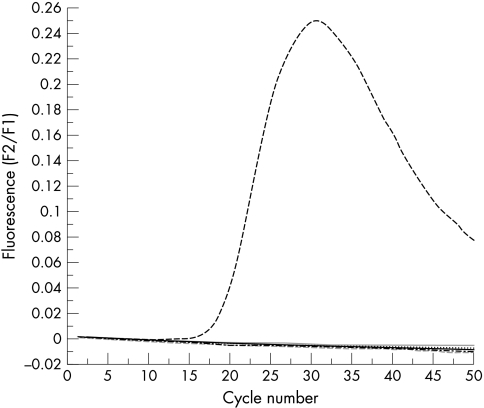
Both real time PCR protocols had the same analytical sensitivity, detecting down to 10−6 pg of SV40 DNA for each reaction, approximately one SV40 copy (fig 1). All of the 56 mesothelioma samples contained amplifiable β globin DNA but none contained amplifiable SV40 DNA with the conventional large T antigen PCR–Southern blotting protocol, protocol 1, or protocol 2. The positive and negative controls gave the expected results (fig 2). There was no evidence of inhibition.
Figure 1.
Real time polymerase chain reaction protocol 1 using serial 10 fold dilutions of control SV40 DNA from 10−4 pg down to 10−8 pg. Amplification occurred at 10−4 pg at 29 cycles, for 10−5 pg at 32 cycles, and for 10−6 pg (approximately one virus copy) at 34 cycles. There was no amplification for 10−7 or 10−8 pg.
Figure 2.
Real time polymerase chain reaction protocol 1 using 30 English mesothelioma samples together with a negative control and a positive control of 1 pg of SV40 DNA. Only the positive control showed amplification.
DISCUSSION
Most studies of SV40 in mesotheliomas have involved mesotheliomas from American patients. Most have demonstrated SV40 DNA in these tumours.2 However, a small minority of recent studies have found no evidence of SV40. It has been suggested that this inconsistency could either be the result of demographic variations in the frequency of SV40 associated mesotheliomas, or differences in methods. The first explanation seems more likely because in some cases the demographic differences have been found to be reproducible from one laboratory to another.4 Emri and colleagues7 could not detect SV40 in 29 Turkish mesotheliomas. De Rienzo and colleagues8 examined 11 US and nine Turkish mesotheliomas in the same laboratory using two primer sets. They found that four of the 11 US mesotheliomas were positive for SV40 with both primer sets but none of the Turkish mesotheliomas was positive. Similarly, Hirvonen and colleagues9 could not detect SV40 DNA in 49 Finnish mesotheliomas, but when the same laboratory examined five mesotheliomas from a New York hospital, as part of a multicentre study,4 they found three to be positive for SV40 DNA.
It has been suggested that the pattern of use of SV40 contaminated polio vaccine could be an explanation for the pronounced differences in the frequency of SV40 detection between countries. Apparently Finland and Turkey did not use contaminated vaccine but the USA did. Widespread polio vaccination in New Zealand started in 1956. By the middle of 1957 approximately 80% of New Zealand children from 5 to 9 years of age had received at least two doses of Salk vaccine imported from the USA.10 In the early 1960s, it was realised that many of these early batches of Salk vaccine from the USA had been contaminated with SV40, although it was not possible retrospectively to determine exactly which batches were infected. This caused some public anxiety at the time, which has been rekindled more recently by the association of SV40 with mesotheliomas. However, our study suggests that this contamination has not given rise to SV40 associated mesotheliomas in New Zealand. Previous studies attempting to demonstrate SV40 virus sequences in mesotheliomas from the UK have yielded variable results. Pepper et al (Cardiff)11 found that using the SV primer set amplification was restricted to four of nine cases of mesothelioma, but six of the nine mesotheliomas showed amplification with the PYV primer set (targeting polyoma virus large T antigen). However, Mulatero et al (London)12 found that 12 of the 17 mesothelioma samples contained amplifiable β globin DNA but none amplified with the PYV primer set.
“It has been suggested that the pattern of use of SV40 contaminated polio vaccine could be an explanation for the pronounced differences in the frequency of SV40 detection between countries”
There is also evidence suggesting that the variations in the reported detection of SV40 may be the result of differences in methods. Strickler et al reported a study in 1996 that did not detect SV40 in 50 US mesotheliomas.13 More recently, Strickler and colleagues14 reported an elaborate multicentre study in which each laboratory received, in a masked fashion, paired replicate DNA samples extracted from 25 fresh frozen mesotheliomas (50 samples) and one from each of 25 normal human lungs. Interspersed were masked positive (titrations of the SV40 genome) and negative control samples. None of the mesotheliomas was found to be reproducibly positive for SV40, despite some laboratories demonstrating sensitivities down to five genome copies in the control specimens.
The third possible explanation for the inconsistency in results between previous studies is that in some studies there has been contamination by SV40. In fact, one study of SV40 in mesotheliomas found that SV40 had contaminated one of the primer sets and one of the negative controls.14 However, some studies have taken elaborate precautions to prevent contamination and have still detected SV40 in mesotheliomas.4 In addition, the detection of SV40 large T antigen in the nuclei of mesothelioma cells cannot easily be explained by contamination.4
In summary, our study describes novel real time FRET probe PCR protocols for the detection of SV40. There is abundant evidence in the literature for the presence of SV40 in mesotheliomas. However, our study shows no evidence of SV40 in mesotheliomas from England and New Zealand. The extensive use of SV40 contaminated polio vaccine in New Zealand does not seem to have resulted in SV40 associated mesotheliomas.
Take home messages.
Despite the fact that there is abundant evidence in the literature for the presence of SV40 in mesotheliomas, we found no evidence of SV40 in mesotheliomas from England and New Zealand
The extensive use of SV40 contaminated polio vaccine in New Zealand does not seem to have resulted in SV40 associated mesotheliomas
Acknowledgments
We thank O Landt (TIB MOLBIOL, Berlin, Germany) for his assistance in the design of the primers and hybridisation probes used in the real time PCR protocols in our study.
Abbreviations
CK, cytokeratin
PCR, polymerase chain reaction
SV40, Simian virus 40
REFERENCES
- 1.Carbone M, Pass HI, Rizzo P, et al. Simian virus 40-like DNA sequences in human pleural mesothelioma. Oncogene 1994;9:1781–90. [PubMed] [Google Scholar]
- 2.Klein G, Powers A, Croce C. Association of SV40 with human tumors. Oncogene 2002;21:1141–9. [DOI] [PubMed] [Google Scholar]
- 3.Mayall FG, Jacobson G, Wilkins R. Mutations of p53 gene and SV40 sequences in asbestos associated and non-asbestos-associated mesotheliomas. J Clin Pathol 1999;52:291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Testa JR, Carbone M, Hirvonen A, et al. A multi-institutional study confirms the presence and expression of simian virus 40 in human malignant mesotheliomas. Cancer Res 1998;58:4505–9. [PubMed] [Google Scholar]
- 5.Pacini F, Vivaldi A, Santoro M, et al. Simian virus 40-like DNA sequences in human papillary thyroid carcinomas. Oncogene 1998;16:665–9. [DOI] [PubMed] [Google Scholar]
- 6.Bergsagel DJ, Finegold MJ, Butel JS, et al. DNA sequences similar to those of simian virus 40 in ependymomas and choroid plexus tumors of childhood. N Engl J Med 1992;326:988–93. [DOI] [PubMed] [Google Scholar]
- 7.Emri S, Kocagoz T, Olut A, et al. Simian virus 40 is not a cofactor in the pathogenesis of environmentally induced malignant pleural mesothelioma in Turkey. Anticancer Res 2000;20:891–4. [PubMed] [Google Scholar]
- 8.De Rienzo A, Tor M, Sterman DH, et al. Detection of SV40 DNA sequences in malignant mesothelioma specimens from the United States, but not from Turkey. J Cell Biochem 2002;84:455–9. [PubMed] [Google Scholar]
- 9.Hirvonen A, Mattson K, Karjalainen A, et al. Simian virus 40 (SV40)-like DNA sequences not detectable in Finnish mesothelioma patients not exposed to SV40-contaminated polio vaccines. Mol Carcinog 1999;26:93–9. [PubMed] [Google Scholar]
- 10.Baguley DM, Glasgow GL. Subacute sclerosing panencephalitis and Salk vaccine. Lancet 1973;2:763–5. [DOI] [PubMed] [Google Scholar]
- 11.Pepper C, Jasani B, Navabi H, et al. Simian virus 40 large T antigen (SV40LTAg) primer specific DNA amplification in human pleural mesothelioma tissue. Thorax 1996;51:1074–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulatero C, Surentheran T, Breuer J, et al. Simian virus 40 and human pleural mesothelioma. Thorax 1999;54:60–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strickler HD, Goedert JJ, Fleming M, et al. Simian virus 40 and pleural mesothelioma in humans. Cancer Epidemiol Biomarkers Prev 1996;5:473–5. [PubMed] [Google Scholar]
- 14.Strickler HD. A multicenter evaluation of assays for detection of SV40 DNA and results in masked mesothelioma specimens. Cancer Epidemiol Biomarkers Prev 2001;10:523–32. [PubMed] [Google Scholar]