Abstract
Aims: To evaluate the IRIS flow microscope for the examination of urine specimens and to compare its performance with that of manual methods; in addition, to assess its usefulness as a screening system for the detection of significant bacteriuria in a routine clinical laboratory.
Methods: The IRIS system was compared with manual quantitative microscopy for the detection of formed elements and with a variety of culture methods to detect urinary tract infection.
Results: The IRIS had a higher sensitivity for the detection of clinically significant formed elements than did manual methods, and using a combination of bacterial and white cell counts produced a sensitivity and specificity of 94.9% and 54.4%, respectively, for predicting positive significant growth, with a 98.5% predictive value for a negative result. Approximately 50% of specimens could be reported as negative on the day of receipt using the screening algorithm.
Conclusions: The sensitivity and specificity of the IRIS is equivalent to that of other automated screening systems and would remove the need for manual microscopy for most specimens. The system was easy to operate and is a useful addition to the methods currently available for the examination of urine specimens.
Keywords: bacteriuria, automation, flow microscopy
Urinary tract infection (UTI) is one of the most common infections diagnosed in clinical microbiology laboratories. Bacteriuria has an important role in disease throughout life. Damage caused by combined reflux and infection is most likely to occur in the first year of life, during the crucial period of renal growth, resulting in cortical damage and scarring, which may lead to renal failure later in life, and 50% of women will have at least one episode of UTI. Urine specimens account for approximately one third of all specimens received in our laboratory, and approximately 80% of these prove to be negative on culture. Numerous methods to screen for bacteriuria have been described previously,1 and automated systems have been developed to screen routine urine specimens, although few have gained widespread acceptance.2 The IRIS 939 UDx (manufactured by International Remote Imaging Systems, Chatsworth, Los Angeles, California and distributed in the UK by Launch Diagnostics Limited, Ash House, New Ash Green, Longfield, Kent) consists of a flow microscope and associated software capable of analysing captured images to identify and quantify formed elements and bacteria in urine.
“Urine specimens account for approximately one third of all specimens received in our laboratory, and approximately 80% of these prove to be negative on culture”
The IRIS system has been available in the USA for several years.3 The earlier versions classified particles by size alone and required the captured images to be verified by the operator. This latest edition has powerful image analysis software that is capable of identifying the following formed elements:
erythrocytes (RBC)
leucocytes (WBC)
leucocyte clumps (WBC clumps)
bacteria (BACT)
yeasts
squamous, transitional, and renal epithelial cells
mucous
sperm
crystals
casts.
METHODS
In total, 905 unselected, consecutive specimens were examined by the IRIS and a comprehensive comparative method. Specimens from all age groups and sexes were included, and all specimens were examined within four hours of receipt in the laboratory.
Sensitivity, specificity, positive, and negative predictive values were calculated using the method of Galen and Gambino.4
Culture comparison
A pour plate viable bacterial counting technique was used as a reference method for the bacterial count. Urine was diluted in quarter strength Ringer’s solution to produce dilutions of 10−2 and 10−4. One millilitre of each dilution and undiluted urine were mixed with 20 ml of sterile, molten Columbia agar base (Oxoid CM311) and plates were incubated overnight at 37°C in air, after which the colonies were counted.
Specimens were also cultured using Bactiuritest strips5 (Mast Group, Bootle, Merseyside, UK) on to CLED medium (Oxoid CM301 Oxoid, Basingstoke, Hampshire, UK) and two Columbia Blood agar (Oxoid CM311) plates incubated in anaerobic and 5% carbon dioxide atmospheres, respectively. Pour plates and Bactiuritest cultures were read independently by two operators. Isolates were identified using a chromogenic medium (CPS IDII; BioMerieux, Basingstoke, Hampshire, UK) or the appropriate API system (BioMerieux).
Each urine was inoculated using a 1 µl loop on to a lawn culture of Bacillus subtilis var globigii and incubated overnight at 37°C. Zones of inhibition around the inoculum were taken to indicate the presence of antimicrobial substances.
Classification of culture positive specimens
A bacterial count of ⩾ 100 000 organisms/ml (with no more than two types of organism), a predominant organism from a mixed culture, or a bacterial count between 10 000 and 100 000 organisms/ml of a pure culture with > 10 WBC/µl in direct microscopy were considered to be positive.
The culture results were classified as shown in table 1.
Table 1.
Classification of culture results
| Reference method | IRIS result | |
| Result | Positive | Negative |
| Positive | True positive | False negative |
| Negative | False positive | True negative |
Microscopy comparison
Duplicate manual microscopic evaluations were performed independently by two operators. Calibrated microscopic examination of uncentrifuged specimens was performed using the Kova® system and inverted microscopy.6 Formed elements were recorded as cells/μl, and the average of the two readings calculated and used for comparison with the IRIS results.
IRIS 939
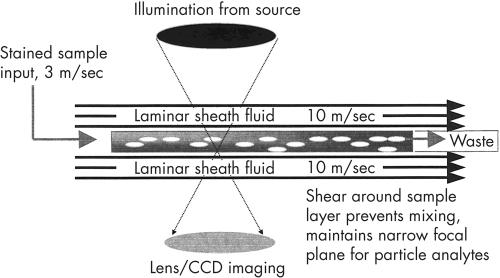
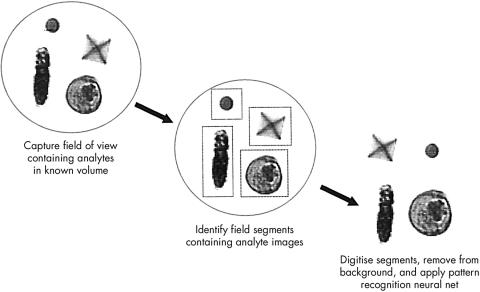
The system consists of two linked units—the flow microscope itself and the view station (a personal computer with high resolution monitor; fig 1). Specimens received in standard plastic 25 ml universal containers are bar coded (using laboratory generated bar codes) with the laboratory number, uncapped, and placed on the instrument carousel, which holds up to 28 specimens. The operator instructs the instrument to begin analysis and the instrument checks the barcode, mixes the specimen, and withdraws 3 ml of sample (sample volumes less than 3 ml require manual microscopy), which is stained and passed to the flow microscope. The system operates by applying hydrodynamic focusing in a sheath fluid; however, unlike most systems using this mechanism, instead of forming a cylindrical sample column the flow cell in the IRIS system shapes the sample into a wide planar stream in the focal plane of the attached microscope (fig 2). An additional effect of this laminar flow is that asymmetric particles align themselves with their narrowest cross section perpendicular to the flow of sheath fluid, allowing the system to differentiate particles in the stream. The flow cell is aligned in the focal plane of the attached microscope. Light is provided by a strobe operating at 30 Hz, and a video camera attached to the microscope is synchronised to the strobe. The captured photographic images are stored and the software identifies “entire” elements, which are digitised and identified by the image analysis program (fig 3). The specimen is first examined under a low power (×10) objective—if no particles are found the system progresses to the next specimen, whereas if there are particles present, the specimen is examined under a high power (×40) objective to enable accurate identification of the particles.
Figure 1.
The IRIS 939Udx instrument, optional strip reader, and view station.
Figure 2.
Diagrammatic representation of hydrodynamic focusing.
Figure 3.
Image analysis process of the IRIS 939 UDx.
Once the system has identified the elements present in the sample, the autoclassification and autorelease programs, in which the user can define their own parameters to select specimens for operator review on the view station, will automatically report specimens that fulfil the user’s criteria. For the purposes of this study, all images were reviewed on the view station and autoclassification and autorelease parameters calculated to give the least number of reviews.
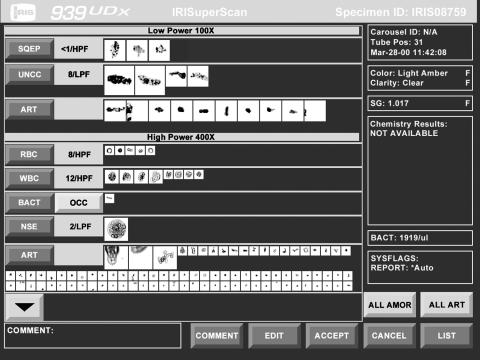
Online review is performed as follows—the instrument displays all captured digital images according to their classification—that is, all RBC images are displayed together, all WBC images together, etc (fig 4). The operator can accept, delete, or reclassify any or all of the displayed images by touching the screen. This process takes only a few seconds and, because of the familiarity of the images, operators are quickly confident in this task.
Figure 4.
An example of the “superscan” editing screen for online review.
Carryover of particles
The system was tested to ensure there was no carryover of particles from one sample to another. This was done in three ways:
Spiking a tube of diluent with a suspension of Escherichia coli and allowing the IRIS to analyse a series of diluent tubes after this.
Spiking sterile urine with a suspension of E coli and allowing the IRIS to analyse a series of diluent tubes after this.
Using a high count clinical sample followed by a series of diluent tubes and allowing the IRIS to process these.
The first two methods were performed in triplicate and the last method was performed in duplicate.
RESULTS
Of the 905 specimens examined, IRIS rejected 40 of these because of “flow errors” (an error code generated when an inadequate volume of specimen passes through the flow cell); this left 865 specimens available for comparison. Of these, 21 had insufficient sample for dilution for pour plate culture and were therefore cultured by bactiuritest.
Formed elements
The Kova system and inverted microscopy were run in parallel for the first 400 specimens. No significant differences in performance were observed between the Kova system and inverted microscopy within operator, so the Kova system was omitted from the remainder of our study and replaced by duplicate inverted microscopy. The results of the manual microscopic investigations were averaged and this value was used in the final comparison; however, the following should be noted:
RBC—paired count coefficients of variation (CVs) for the duplicate counts averaged over 34% and there were 30 samples that had one high count (⩾ 50) and one negative count (0).
WBC—paired count CVs for the duplicate counts averaged 28% and there were 13 samples that had one high count (⩾ 50) and one negative count (0).
Squamous epithelial cells (SEC)—paired count CVs for the duplicate counts averaged 32% and there were 58 samples where one value was high and the other zero.
Tables 2–4 show a comparison of the IRIS results with manual microscopy for the above elements. As outlined above, all of the IRIS images were reviewed online by two independent operators and edited; these results for WBC and RBC are also shown in tables 2–4.
Table 2.
Leucocyte counts: IRIS compared with manual microscopy before and after review
| Manual count (cells/µl) | IRIS count (cells/µl) | |||||||
| <10 | 10–49 | 50–99 | ⩾100 | |||||
| Pre | Post | Pre | Post | Pre | Post | Pre | Post | |
| <10 | 245 | 327 | 132 | 88 | 27 | 12 | 34 | 11 |
| 10–49 | 43 | 82 | 80 | 61 | 34 | 27 | 30 | 18 |
| 50–99 | 4 | 8 | 10 | 11 | 17 | 16 | 26 | 22 |
| ⩾100 | 1 | 2 | 5 | 7 | 15 | 21 | 162 | 152 |
Table 4.
Epithelial cells: IRIS compared with microscopy
| Manual count (cells/µl) | IRIS count (cells/µl) | |||
| <10 | 10–49 | 50–99 | ⩾100 | |
| <10 | 540 | 23 | 0 | 0 |
| 10–49 | 137 | 75 | 4 | 0 |
| 50–99 | 6 | 35 | 8 | 1 |
| ⩾100 | 3 | 7 | 20 | 6 |
Table 2 shows the results for WBC—there was absolute agreement (using the groupings shown) of 58%; from table 2 it can be seen that IRIS missed only one significant WBC count. Following review, the agreement rate was 64%, although there were then two specimens where IRIS failed to show significant WBC. Table 3 shows the results for RBC—once again, the absolute agreement was good at 63%, with IRIS missing only three significant RBC counts. After review, the agreement again rose to 80% and there were six specimens where IRIS failed to show significant RBC. Table 4 shows the results for SEC—absolute agreement was good at 73%, with IRIS missing only three specimens with significant SEC by manual microscopy.
Table 3.
Erythrocyte counts: IRIS compared with manual microscopy before and after review
| Manual count (cells/µl) | IRIS count (cells/µl) | |||||||
| <10 | 10–49 | 50–99 | ⩾100 | |||||
| Pre | Post | Pre | Post | Pre | Post | Pre | Post | |
| <10 | 430 | 589 | 119 | 31 | 36 | 6 | 50 | 9 |
| 10–49 | 36 | 70 | 44 | 36 | 16 | 7 | 22 | 5 |
| 50–99 | 7 | 11 | 9 | 7 | 6 | 3 | 7 | 8 |
| ⩾100 | 3 | 6 | 5 | 5 | 10 | 7 | 65 | 65 |
Casts—manual microscopy detected 33 specimens with casts; IRIS failed to detect 13 of these. IRIS flagged 268 specimens as containing casts but upon online review only 37 were found to contain images of true casts; manual microscopy missed 20 of these. There was agreement in only 17 specimens.
Crystals—the IRIS autoclassification system reports total crystals and produced a significant number of false positives, with only 29 of the 192 flagged specimens being found positive upon review.
Culture
Three hundred and eighty four of the samples had pour plate counts of > 100 000 organisms/ml and 366 had Bactiuritest counts of >100 000 organisms/ml. Of these 384, 136 specimens were classified as culture positive by the criteria outlined above, 130 by the pour plate method, and 136 by Bactiuritest.
Ninety one specimens demonstrated the presence of antimicrobial substances, 14 of which were in the culture positive group.
Table 5 shows the performance of the IRIS bacterial count compared with the viable count. Using the BACT count alone for determining significant bacteriuria, a sensitivity of 84.6% and a specificity of 65.0% was the best that could be achieved. However, by combining the BACT count and WBC count and applying iterative testing to various combinations (table 6) a combination of ⩾ 15 BACT or ⩾ 80 WBC produced a sensitivity and specificity of 94.9% and 54.4%, respectively. Using these values for detecting all bacterial growth of ⩾ 100 000 organisms/ml the system had a sensitivity and specificity of 82.5% and 67.5%, respectively. Furthermore, using an algorithm set at < 15 BACT/µl and < 80 WBC/µl gives a negative predictive value of 98.5%, with a positive predictive value of 28%, rejecting 392 negative specimens. This leaves a total of 473 specimens that would have been flagged for culture, resulting in 343 false positives and seven false negatives, five of which had no evidence of pyuria and were therefore thought to be of doubtful relevance.
Table 5.
IRIS BACT count compared with viable count
| Viable count (cfu/ml) | IRIS BACT count (cells/µl) | |||
| <1 | 1–9 | 10–99 | ⩾100 | |
| <1000 | 281 | 41 | 46 | 28 |
| ⩾1000 to <10 000 | 5 | 0 | 0 | 0 |
| ⩾10 000 to <100 000 | 53 | 5 | 16 | 6 |
| ⩾100 000 | 76 | 24 | 112 | 172 |
cfu, colony forming units.
Table 6.
The performance of IRIS evaluated by iterative testing using various thresholds
| Criteria – bacteria and WBC count | Neg. Crit. + Neg. Cult. | Pos. Crit. + Pos. Cult. | Pos. Crit. + Neg. Cult. | Neg. Crit. + Pos. Cult. | Specificity | Sensitivity |
| 939 ⩾15 BACT or ⩾100 WBC | 394 | 129 | 334 | 8 | 54.1% | 94.8% |
| 939 ⩾15 BACT or ⩾80 WBC | 385 | 130 | 343 | 7 | 54.4% | 94.9% |
| 939 ⩾15 BACT or ⩾50 WBC | 352 | 131 | 376 | 6 | 48.4% | 95.6% |
BACT, bacteria; Neg. Crit., negative criteria; Neg. Cult., negative culture; Pos. Crit., positive criteria; Pos. Cult., positive culture; WBC, leucocytes.
Carryover tests
To determine the efficacy of the probe washing system the procedures described above were performed. After analysis, all tubes were subcultured using a Bactiuritest strip. When using a culture positive urine or spiked sterile urine as the inoculum no detectable carryover was demonstrated. However, using saline spiked with > 100 000 colony forming units (cfu)/ml of E coli there was detectable carryover into the first diluent tube.
DISCUSSION
Our results show that the system could reliably detect and classify the major formed elements found in clinical specimens. It was interesting to note that reviewing the specimens had little effect on the significance of most of the results. Indeed, after review, the IRIS proved more sensitive for the detection of WBC and RBC, probably as a result of the increased sample volume examined. However, it of interest that there was an increase in “false negatives” after review, because clearly the reviewers did see images of these elements, and it may be that the manual microscopy was in fact a false positive. It was of course not possible to re-examine the original specimens after result analysis, although IRIS images are still available for examination. Overall, the system proved more sensitive for the detection of clinically important elements, but less sensitive than manual systems for crystals. In the case of casts, neither the IRIS nor manual system showed an advantage in performance, reflecting the difficulty in looking for these elements in a single specimen. It seems clear from these data that the IRIS would be a suitable method for the routine examination of clinical specimens for formed elements, and would allow the de-skilling of this time consuming task.
It was interesting to note that most “one replicate positive the other negative” errors in manual counts occurred when there were very high counts of a particular element. The same was found, but to a lesser extent, when reviewing IRIS results—most of the counts that were altered were the result of incorrect classification of some particles with high count specimens.
“The system proved more sensitive for the detection of clinically important elements, but less sensitive than manual systems for crystals”
Because of the clarity of the images and the way in which the screens are organised reviewing is a very rapid process, being just a matter of a few seconds for each specimen. When alterations are required the touch screen makes this process very efficient.
Although the Bactiuritest method appears to have detected all the classified positive specimens, five of the six specimens had insufficient sample for pour plate culture and the remaining sample had a calculated viable count of 50 000 cfu/ml.
Although the IRIS BACT count was less sensitive and specific than viable counting methods for the detection of bacteria, when combined with the results of WBC counts a sensitive result was achieved. Using the algorithm < 15 BACT/µl and < 80 WBC/µl the system had a predictive negative value of 98.7% and rejected 45% of the specimens examined, with only seven false negatives, five of which had no evidence of pyuria and were therefore thought to be of doubtful relevance. However, it should be noted that of the 343 specimens classified as false positives, 169 had bacterial counts > 100 000/ml, 53 specimens had counts in excess of 100 000/ml of carboxyphilic organisms, and a further 29 proved positive for antimicrobial substances. It was interesting to note that following review of the results the performance of this algorithm worsened, and it would seem advisable to screen the specimens using the IRIS result and only edit the review specimens after this process has been completed. It may be possible to refine this algorithm further because it is possible to set different interpretation criteria for particular patient groups (for example, males and females); however, this was outside the remit of our study.
These results compare well with other published data using automated urine screening systems,2,7 where sensitivities of 93% and specificities of 74% were reported.
Carryover was the only concern that we had about the performance of the system. Although statistically not important to the performance of the system, the carryover of viable organisms and formed elements to subsequent specimens could have serious consequences on the result of a particular specimen. It should be emphasised that in the USA separate specimens are requested for culture examination if screening tests suggest infection, so that this phenomenon is not of great importance. After discussions with the manufacturer, a modification to the system has been made, which effectively sterilises and washes the liquid handling system in between specimens, thereby eliminating carryover of particles. This is carried out by rinsing the sampling probe in a diluted proprietary disinfectant and then rinsing (personal communication, W M Dougherty, 2002).
The IRIS system is very easy to operate. It has several internal checks to ensure optimum performance and reproducibility of the system, and presents its results in a format that is familiar to microbiologists. In this evaluation, it was possible to process approximately 40–50 specimens each hour on IRIS. This process could easily be performed by a medical laboratory assistant, and the complete final results of flagged negative specimens and microscopic examinations of flagged positive specimens would be available on the day of receipt. It is also possible to interrupt the system to perform urgent examinations as a STAT procedure. Using the optimised autoclassification and autorelease thresholds from this study, it was calculated that the on screen review rate for these specimens would be 20%.
Take home messages.
The sensitivity and specificity of the IRIS is equivalent to that of other automated screening systems and would remove the need for manual microscopy for most specimens
It has a rapid turnaround time, which could prove beneficial for patient care
The system was easy to operate and could be used with confidence by any laboratory scientist
The system is a useful addition to the methods currently available for the examination of urine specimens
“Carryover was the only concern that we had about the performance of the system”
This rapid turnaround time could prove beneficial for patient care, but does depend to some extent on the speed of transport of specimens to the laboratory and the method by which results are returned to requesting clinicians. The ability to interface the instrument to the laboratory’s host system has great advantages, not only in terms of speed of reporting, but also data integrity. The reduction in the number of specimens requiring culture and therefore interpretation the following day would also have definite advantages in terms of work flow for the laboratory. The only limitation of the system is the minimum sample volume of 3 ml, although a paediatric protocol allowing the use of smaller volumes is available.
The list price capital cost of the instrument is £72 000, and the consumable costs are not yet finalised, but will be dependant upon the number of specimens analysed.
Our evaluation was the first time that this instrument has been used as a stand alone system for the routine screening of clinical specimens for evidence of UTI.
The system was simple to operate and had clear and easily understood software. The images for review were readily recognisable and any laboratory scientist would be confident in performing this task. The ability of the system to use primary tube sampling and to provide a rapid and sensitive result makes this a useful system for screening for bacteriuria.
Acknowledgments
The authors wish to acknowledge the expert technical assistance of Miss D Wadham, and Launch Diagnostics for providing the IRIS instrument.
Abbreviations
BACT, bacteria
cfu, colony forming units
RBC, erythrocytes
SEC, squamous epithelial cells
UTI, urinary tract infection
WBC, leucocytes
REFERENCES
- 1.Stevens M. Screening urines for bacteriuria. Med Lab Sci 1989;46:194–206. [PubMed] [Google Scholar]
- 2.Stevens M, Mitchell CJ, Livsey SA, et al. Evaluation of Questor urine screening system for bacteriuria and pyuria. J Clin Pathol 1993;46:817–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roe CE, Carlson DA, Daigneault RW, et al. Evaluation of the Yellow IRIS. Am J Clin Pathol 1986;86:661–5. [DOI] [PubMed] [Google Scholar]
- 4.Galen RS, Gambino SR. Beyond normality: the predicative value and efficiency of medical diagnoses. New York: John Wiley and Sons, 1975.
- 5.Leigh DA, Williams JD. Method for the detection of significant bacteriuria in large groups of patients. J Clin Pathol 1964;17:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maskell R. Diagnosis of urinary tract infection, its causes and consequences. In: Urinary tract infection. Current Topics in Infection Series 3. London: Edward Arnold: 21–41.
- 7.Toffaletti J, Dotson MA, Shearman P, et al. Comparison of 2 automated systems for urine chemistry and urine sediment analysis. Lab Haematol 1999;5:123–9. [Google Scholar]