Abstract
Aims: To study the association between simian virus 40 (SV40) and human hepatocarcinogenesis.
Methods: Polymerase chain reaction (PCR) to detect SV40 large T antigen (Tag) DNA was performed on: 50 human hepatocellular carcinoma (HCCs) diagnosed between 1978 and 1989 (cohort A); 20 cases of alcoholic liver cirrhosis from the same period; and 20 HCCs diagnosed after 1997 (cohort B). PCR to detect SV40 regulatory sequence and SV40 Tag immunohistochemistry were performed on selected cases from cohorts A and B. Amplified products were directly sequenced. Immunohistochemistry for p53 and pRb and clinicopathological analyses were performed on selected cases from cohorts A and B. Complete survival data were collected for cohort A.
Result: SV40 Tag DNA was found in five cohort A HCCs but not in alcoholic liver cirrhosis cases or cohort B HCCs. Neither SV40 regulatory sequence nor SV40 Tag protein were demonstrated in Tag DNA positive HCCs. No clinicopathological differences existed between Tag DNA positive and negative HCCs, but the presence of Tag DNA was associated with reduced disease specific survival. Relatively fewer Tag DNA positive than negative HCCs expressed p53, but loss of pRb expression was similar in the two groups. Patients with Tag DNA positive HCCs were unlikely to have received SV40 contaminated poliovirus vaccine.
Conclusions: SV40 Tag DNA is present in a small proportion of historical HCCs and may contribute to their pathogenesis and influence their outcome. The source of the virus is uncertain and more recent HCCs show no evidence of SV40.
Keywords: Simian virus 40, hepatocellular carcinoma, prognosis, p53, retinoblastoma protein
Recent figures show that hepatocellular carcinoma (HCC) is the third and fifth most common cause of cancer death among men and women, respectively, worldwide.1 Several aetiological factors are already known, including glycogen storage diseases, haemochromatosis, aflatoxin exposure, and hepatitis B or C viral infection.2 However, identifying new risk factors for HCC remains important, because it may contribute to the identification of patients at high risk of developing the cancer, and provide further insight into the molecular mechanisms that underlie hepatocarcinogenesis.
“We looked for simian virus 40 DNA sequences and large T antigen protein in well defined cohorts of human hepatocellular carcinomas”
Simian virus 40 (SV40) is a small, double stranded DNA virus that was discovered as a contaminant of inactivated polio vaccines.3,4 Although predominantly thought of as a simian virus, SV40 has been demonstrated, with replicative ability, within the human population.3,4 The introduction of SV40 into humans may relate to the widespread use of SV40 contaminated inactivated polio vaccine in several Western countries, including the USA and the UK in the 1950s and 1960s.3,5 Furthermore, it has been suggested that the virus may be maintained in the human population by horizontal and vertical transmission.6,7 The relevance of SV40 to humans relates to its possible aetiological role in several human tumours. The presence of SV40 DNA and its large T antigen (Tag) was first demonstrated by PCR analysis in choroid plexus tumours and ependymomas.8 Since then, however, SV40 DNA has also been isolated from several other human neoplasms including osteosarcoma,6 mesothelioma,9 papillary thyroid carcinoma,10 and lymphoma.7,11,12 Several factors point towards a possible, similar link between SV40 and HCC. First, several breeds of transgenic mice engineered to express the SV40 Tag within the liver show spontaneous development of HCC13–15; indeed, the occurrence of such tumours is inevitable in at least one breed.13 Second, when combined with mutation of the c-H-ras oncogene, transfection with SV40 Tag leads to full transformation of hepatocytes in vitro.16 Finally, SV40 Tag has been shown to inactivate both p53 and retinoblastoma protein (pRb),17 and loss of function of these two oncosuppressor proteins appears to be important to the process of hepatocarcinogenesis—genetic abnormalities of p53 and Rb have been recorded in as many as 60% and 35% of human HCCs, respectively.18,19 Despite these suggestions of a possible link between SV40 and human HCC, we are aware of only one previous attempt to test this hypothesis. Lednicky and Butel reported that no SV40 DNA was isolated from “a limited number” of HCCs, but neither the number of cases studied nor the experimental or clinicopathological details were given.20
In our present study, we looked for SV40 DNA sequences and Tag protein in well defined cohorts of human HCCs. We also investigated whether the presence of SV40 DNA was correlated with any clinicopathological factor, including prognosis and likely exposure to SV40 contaminated inactivated polio vaccine.
METHODS
Cases and survival analysis
Two cohorts of HCCs were studied. The first (cohort A) comprised 50 cases diagnosed between 1978 and 1989, and the second (cohort B) comprised 20 cases diagnosed between 1998 and 2001. Twenty cases of alcohol related cirrhosis diagnosed between 1978 and 1989 were also studied. Paraffin wax embedded, formalin fixed tissue blocks from all the above cases (tumour tissue for HCC cases and cirrhotic liver tissue for the alcohol related cirrhosis cases) were retrieved from the archival files of the department of pathology at the University of Edinburgh Medical School, UK. Haematoxylin and eosin stained sections of the carcinomas were reviewed by a gastrointestinal histopathologist (NW), taking into account the degree of differentiation (using the four point scale of Edmondson and Steiner),2 the main cell types (for example, hepatoid and/or clear), and the predominant growth pattern (trabecular, acinar, and/or solid). Date of birth, sex, and presence and type of any background liver disease were also compiled for each patient with HCC. All 70 patients with HCC had been managed at the Royal Infirmary of Edinburgh, UK. Patient details for cohort A were given to the Scottish Cancer Registry to enable a search for dates and registered causes of deaths. For any patients currently living at the time of our study, a survival time was calculated using 1 February 2003 as the census point. Our study was carried out in accordance with the guidelines of the Lothian local research ethics committee.
DNA analyses
DNA was extracted from sections cut from the tissue blocks of the 70 cases of HCC and 20 cases of alcohol related cirrhosis, using xylene dissolution, proteinase K digestion, and lysis, followed by ethanol precipitation. Complete details of the DNA extraction technique are provided elsewhere.21 The integrity of the extracted DNA had been confirmed from a previous study of the samples.22,23 Briefly, PCRs for several genes (for example, β actin and microsomal epoxide hydrolase, producing intact fragments of up to 250 bp) were consistently successful for all samples studied. Furthermore, spectrophotometric readings of the extracted DNA tallied with quantities suggested by agarose gel electrophoresis run against known standards.
The primers SV.for3 (5′-TGA GGC TAC TGC TGA CTC TCA ACA-3′) and SV.rev (5′-GCA TGA CTC CAA AAC TTA GCA ATT CTG-3′) amplify a Tag specific sequence adjacent to the pRb binding site,8 and are particularly suited for use on DNA extracted from paraffin wax embedded tissues.24 The presence of the SV40 Tag DNA sequence was further investigated using a second PCR with the Pyv set of primers (Pyv.for, 5′-TAG GTG CCA ACC TAT GGA ACA GA-3′; Pyv.rev, 5′-GGA AAG TCT TTA GGG TCT TCT ACC-3′), which overlap the SV.for3/rev primer pair.8 For both the SV.for3/rev and the Pyv PCRs, a 50 µl reaction volume was used, comprising final quantities/concentrations of 100 ng of template DNA, 0.2 μM of each primer, 1 U of Taq polymerase, 1.5 mM of MgCl2, and 200 μM of each dNTP. The cycling conditions used were a denaturation step of 94°C for 12 minutes, 45 cycles of 94°C for 45 seconds, 55°C (SV.for3/rev) or 53°C (Pyv) for 60 seconds, and 72°C for 45 seconds, followed by a final extension step of 72°C for 10 minutes. A plasmid (pRen2) containing the SV40 Tag sequence (gift from Professor JJ Mullins, School of Clinical Sciences and Community Health, University of Edinburgh, UK) was used as a positive control for the SV.for3/rev and Pyv PCRs.
Tag sequences are also shared by related viruses such as the JC and BK viruses.3,4 Therefore, to be certain that SV40 DNA is specifically being detected, previous investigators have, in addition to Tag targeted PCR: (1) used additional sets of primers that amplify other sequences of the SV40 genome10,24; (2) performed Southern blotting with specific probes for SV40, JC, and BK viruses8,10; and/or (3) sequenced the products of the Tag targeted PCR.11,12 The primers RA1 (5′-AAT GTG TGT CAG TTA GGG TGT-3′) and RA2 (5′-TCC AAA AAA GCC TCC TCA CTA CTT-3′) target the SV40 regulatory region and span an area showing 72 bp insert repeats, thus allowing for the distinction between laboratory and “natural” strains of the virus.24,25 For this PCR, a 50 μl reaction volume was used, comprising final quantities/concentrations of 100 ng of template DNA, 0.2 μM of each primer, 1 U of Taq polymerase, 1.5 mM of MgCl2, and 200 μM of each dNTP. The cycling conditions used were a denaturation step of 94°C for 12 minutes, 45 cycles of 94°C for 60 seconds, 55°C for 60 seconds, and 72°C for 60 seconds, with a final extension step of 72°C for 10 minutes. The pcDNA3.1 plasmid (Invitrogen Ltd, Paisley, UK) contains the SV40 regulatory sequence and was used as a positive control for the RA1/2 PCR.
The sensitivities of the SV.for3/rev, Pyv, and RA1/2 PCRs were assessed using reconstruction experiments. Each experiment used 100 ng aliquots of human liver genomic DNA spiked with serial tenfold dilutions (equivalent to 100 ng to 1 ag) of the respective positive control plasmid.
The PCR products were run on 2% agarose electrophoresis gels and visualised using ethidium bromide. Any amplified products were directly sequenced (DNASHEF Technologies, Edinburgh, UK) using a fluorescent sequencer (ABI 377). The amplified sequences were compared against the known SV40 genome using the Genebank/EMBL database (via the NCBI Blast website: http://www.ncbi.nlm.nih.gov/blast/).
Immunohistochemistry
We are aware of only one antibody that binds to SV40 Tag but not SV40 small t antigen or JC or BK Tag.26 However, the sensitivity of this antibody (Pab 101) is compromised by the fact that it is targeted at the C-terminus of the Tag protein, which has been shown to contain a variable region.27 Therefore, to optimise the sensitivity of the immunohistochemical detection of Tag, previous investigators have used an additional antibody (Pab 419), which binds to the better conserved N-terminus of the SV40 Tag protein, but also crossreacts with SV40 small t antigen, although not with JC or BK Tag.28
For immunohistochemistry, 4 μm tissue sections were cut, dewaxed, and rehydrated. For Pab 101 (Santa Cruz Biotechnology via Insight Biotechnology, Wembley, Middlesex, UK) and Pab 419 (Oncogene Research via Merck Biosciences, Nottingham, UK) immunohistochemistry, sections were subjected to pressure cooking antigen retrieval for three minutes in citrate buffer (10mM, pH 6.0). Endogenous peroxidase was blocked with 1.5% H202 in methanol and the sections then incubated in primary antibody at a 1/50 dilution for one hour. For p53, sections were subjected to microwave antigen retrieval for 10 minutes in citrate buffer (10mM, pH 6.0). Endogenous peroxidase was blocked with 1.5% H202 in methanol and the sections were then incubated in primary antibody (clone DO7; Dako, Ely, Cambridgeshire, UK) at a 1/200 dilution for 30 minutes. For pRb, sections were subjected to microwave antigen retrieval for 10 minutes in citrate buffer (10mM, pH 6.0). Endogenous peroxidase was blocked with 1.5% H202 in methanol and the sections were then incubated in primary antibody (clone Rb1; Novocastra, Newcastle upon Tyne, UK), at a 1/25 dilution at 4°C overnight. For all four antibodies, biotinylated rabbit antimouse secondary antibody (Dako) was used at a 1/400 dilution for 30 minutes, and detection was carried out using commercial avidin–biotin complex (Vector Laboratories, Peterborough, UK; 30 minutes) and diaminobenzidine reagents (Dako; five minutes). Mayer’s haematoxylin was used as a counterstain for visualisation.
Formalin fixed cell blocks prepared from a neuronal cell line established from a transgenic SV40 Tag mouse (gift from Dr S Wharton, Department of Pathology, University of Edinburgh, UK) were used as positive controls for both Pab 101 and Pab 419 immunohistochemistry. A colorectal carcinoma with a known p53 mutation and stabilised p53 protein was used as a positive control for p53 immunohistochemistry, and normal tonsil was used as a positive control for pRb immunohistochemistry. Endothelial cells also served as internal positive controls for pRb immunohistochemistry.
Statistical analyses
Categorical data were analysed using a two tailed χ2 test (with Yates’ correction for 2 × 2 table analysis) or, where more appropriate, a two tailed Fisher’s exact test. Univariate survival analysis was carried out using Kaplan-Meier plots and the log rank test.
RESULTS
The SV.for3/rev, Pyv, and RA1/2 PCR protocols used were able to detect, on a background of 100 ng of genomic human DNA, up to 100 ag, 1 fg, and 1 fg of positive control plasmid DNA, respectively. Taking an approximate DNA content of a single human cell as 6 pg and the size ratios of each PCR amplicon (see below) to the respective positive control plasmids (pRen2/Tag, 10.6 kb; pcDNA3.1, 5.4 kb) suggests that the SV.for3/rev, Pyv, and RA1/2 PCRs could detect at least 9, 86, and 169 copies, respectively, of target sequence among 100 ng of sample DNA (approximately equivalent to 16 667 cells).
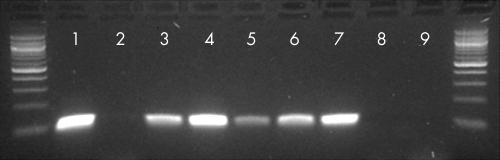
All 70 HCC cases and 20 alcohol related cirrhosis cases were studied with the SV.for3/rev primers. Of these cases, only five (all carcinomas from cohort A) yielded a single product of equal size (105 bp) to the positive control (fig 1). In all five positive cases, sequencing and analysis of the amplified product showed complete homology with an SV40 Tag specific sequence (adjacent to the pRb binding site). Three of these five cases showed products of the expected size (172 bp) with the Pyv PCR. However, none of the five cases or a further 15 cohort A HCCs studied yielded products (expected size 242 bp) with the RA1/2 PCR. None of the five cases positive for the SV40 Tag DNA sequence and none of a further 20 cohort A HCCs or 20 cohort B HCCs studied showed immunoreactivity for either Tag antibody.
Figure 1.
Amplification products from polymerase chain reaction using SV.for3/rev primers; 100 bp ladders are shown on either side of the gel. Lanes 1 and 2 represent positive and water blank controls, respectively. Lanes 3 to 7 represent five cases of hepatocellular carcinoma from which simian virus 40 (SV40) large T antigen (Tag) DNA sequences could be amplified. Lanes 8 and 9 represent two examples of hepatocellular carcinoma (from cohort A) from which SV40 Tag DNA sequences could not be amplified.
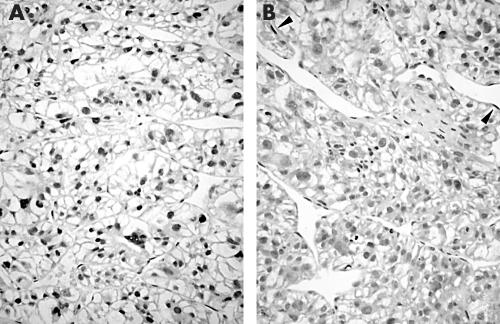
Table 1 shows the clinicopathological details of the five patients with HCC who were positive for the SV40 Tag DNA sequence. Although the group sizes were small, there were no significant differences in these details between the five patients and a further 20 cohort A HCCs studied or the 20 cohort B HCCs (table 1; statistical analyses not shown but available on request). Only one of the five cases positive for the SV40 Tag DNA sequence showed p53 immunoreactivity (fig 2A), compared with 10 of the further 20 cohort A HCCs studied (two tailed Fisher’s exact test; p = 0.34). Three of the five cases positive for SV40 Tag DNA sequence lacked pRb immunoreactivity (fig 2B) compared with nine of a further 20 cohort A HCCs studied (two tailed Fisher’s exact test; p = 0.64).
Table 1.
Clinicopathological features of the simian virus 40 (SV40) large T antigen sequence positive hepatocellular carcinomas and 20 carcinomas each from cohorts A and B
| Clinicopathological feature | SV40+ve cohort A cases (n = 5) | SV40–ve cohort A cases (n = 20) | Cohort B cases (n = 20) |
| Age (years) | |||
| Median | 68 | 66.5 | 57 |
| Range | 45–82 | 22–88 | 42–75 |
| Sex | |||
| Male | 4 | 14 | 14 |
| Female | 1 | 6 | 6 |
| Background cirrhosis | |||
| Yes | 4 | 13 | 16 |
| No | 1 | 7 | 4 |
| Tumour grade* | |||
| I/II | 2 | 6 | 9 |
| III/IV | 3 | 14 | 11 |
| Tumour cell type | |||
| Hepatoid only | 4 | 13 | 13 |
| Hepatoid+clear | 1 | 7 | 7 |
| Predominant tumour architecture | |||
| Trabecular | 2 | 12 | 12 |
| Solid | 2 | 3 | 6 |
| Acinar | 1 | 5 | 2 |
*Based on the classification of Edmondson and Steiner.2
Figure 2.
Example of a hepatocellular carcinoma immunostained for (A) p53 and (B) retinoblastoma protein (pRb). There is strong diffuse nuclear staining for p53, suggesting the presence of gene mutation. There is also loss of pRb expression compared with endothelial cells (arrow heads), which act as an internal positive control.
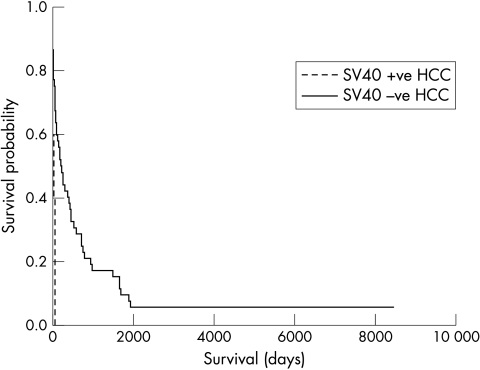
The median follow up period of the cohort A patients was 249 months (range, 163–302). Forty seven of these 50 patients had died by the time of census. All but two of these deaths had HCC as the primary registered cause of death. The five patients with SV40 Tag DNA sequence positive HCCs showed a shorter median disease specific survival compared with the remaining 45 patients from cohort A (41 versus 207 days, respectively; log rank test, p = 0.0021; fig 3).
Figure 3.
Kaplan-Meier plot showing outcome of cohort A patients divided into those with and without simian virus 40 (SV40) large T antigen DNA sequence positive hepatocellular carcinomas (HCCs). Log rank test, p = 0.0021.
DISCUSSION
To the best of our knowledge, this is the first extensive search for SV40 in human HCC. We successfully amplified SV40 Tag DNA sequences from five of 50 historical HCCs. It is conceivable, however, that this represents an underestimate. Our SV.for3/rev PCR should have been able to detect much less than one copy of SV40 DNA for each tumour cell, but the DNA ethanol precipitation method used may not have recovered minute amounts of viral DNA in the first place. The fact that neither the SV40 regulatory sequence nor the Tag protein could be demonstrated in the five SV40 Tag DNA positive HCCs also needs further discussion. The possibility of contamination was considered because work on SV40 Tag transgenic murine tissue had previously been carried out in the laboratory where our PCR experiments were performed. However, when DNA was re-extracted from different tissue blocks from three of the five SV40 Tag DNA positive HCCs and amplified in a different laboratory, the same sequences could still be demonstrated. It is feasible that the immunohistochemical protocols used were not sufficiently sensitive to detect the small amounts of SV40 Tag protein present in the HCCs compared with the positive control cells. Furthermore, whereas 100% of hepatocytes showed immunohistochemical evidence of Tag protein in the early stages of hepatocarcinogenesis in a transgenic SV40 Tag mouse, some tumour nodules subsequently lost complete expression of the protein.29 There are two possible reasons why we failed to demonstrate SV40 regulatory sequences despite the presence of Tag DNA sequences; that is, failure of the RA1/2 PCR (because of DNA of insufficient quality and/or inadequate PCR sensitivity) or the absence of the complete SV40 genome in the Tag DNA positive HCCs. In previous studies of the same DNA samples, we had consistently amplified fragments of up to 250 bp, and the expected product (242 bp) from the RA1/2 PCR lies within this size range. Nonetheless, this larger size (compared with the 105 bp amplicon of the SV.for3/rev PCR), together with a lower assay sensitivity, may have been sufficient to yield a negative RA1/2 PCR, despite a positive SV.for3/rev PCR. In support of this, the Pyv PCR also showed a lower sensitivity than the SV.for3/rev PCR, and was only able to confirm the presence of the SV40 Tag DNA sequence in three of the five HCCs that were positive with the SV.for3/rev PCR. With regard to the alternative explanation for failure to detect SV40 regulatory sequences, the probable presence of only subgenomic portions of SV40 has previously been reported in other tumours.25 However, if truly present, the relevance of incomplete SV40 genomes to human neoplasia remains uncertain. Regardless of the reason for demonstrating only SV40 Tag DNA sequences, it was still of interest to determine whether their presence might be related to the pathogenesis and/or outcome of human HCCs.
“Our sample size of large T antigen (Tag) DNA sequence positive hepatocellular carcinomas was small, but there was a suggestion that these cancers less frequently showed p53 protein expression (and by inference p53 mutation) than their Tag negative counterparts”
SV40 Tag can inhibit both p53 and pRb activity,17 and loss of both oncosuppressor proteins (usually through somatic mutation or gene deletion) is important to the process of hepatocarcinogenesis.18,19 If SV40 infection plays a role in the pathogenesis of a proportion of human HCCs, it is possible that SV40 Tag could substitute for the need for p53 and pRb genetic abnormalities in these cancers. Indeed, liver tumours arising in transgenic SV40 rodents show no evidence of mutant p53.30 Unfortunately, the historical nature of the Tag DNA positive HCCs meant their extracted DNA was of insufficient quality to permit informative analyses of p53 and Rb. Therefore, we used p53 and pRb immunohistochemistry to reflect abnormalities of their respective genes. Immunohistochemical detection of p53 may serve as a surrogate (albeit not absolute) marker for gene mutation,31 whereas loss of pRb expression correlates well with Rb mutation and/or loss of heterozygosity (LOH) within HCCs.32 Our sample size of Tag DNA sequence positive HCCs was small, but there was a suggestion that these cancers less frequently showed p53 protein expression (and by inference p53 mutation) than their Tag negative counterparts. Mayall and colleagues similarly found that one of five SV40 Tag DNA sequence positive mesotheliomas showed p53 immunostaining, compared with four of six Tag negative mesotheliomas.33 There have been few formal attempts to clarify whether the presence of SV40 Tag can truly negate the need for p53 genetic abnormalities in human malignancies in vivo. However, complexes of p53 and SV40 Tag protein have been demonstrated in several such malignancies,34,35 and among neoplasms arising on a background of Li-Fraumeni syndrome, SV40 Tag has been found only in tumours retaining the normal p53 allele, and not in those showing LOH of p53.36 Unlike p53, there was no obvious relation between the presence of the SV40 Tag DNA sequence and pRb expression among our HCCs. Therefore, it is possible at most, that SV40 Tag contributes to human hepatocarcinogenesis by substituting for p53 genetic aberrations. However, it is uncertain whether SV40 infection alone would be sufficient to induce and drive the whole process of hepatocarcinogenesis in humans. Although some SV40 transgenic mouse models spontaneously develop hepatomas,13 ras mutation is required in addition to SV40 Tag transfection for full transformation of cultured hepatocytes.16
Regardless of whether SV40 infection can truly promote hepatocarcinogenesis, the presence of the virus or its Tag sequence could still theoretically influence phenotype and/or outcome of the malignancy. We found no clinicopathological differences between SV40 Tag DNA positive and negative HCCs. However, the presence of the Tag DNA sequence was associated with a significant reduction in median disease specific survival. All our cases of HCC had been managed in the same hospital and our survival analyses were only applied to cases from the same time period to minimise bias from temporal changes in clinical management. There have been few formal attempts to test whether the presence of SV40 Tag confers a worse prognosis for a human malignancy. However, in a recent study of 83 mesotheliomas, Procopio and colleagues also demonstrated a trend towards shorter survival among patients with SV40 positive tumours compared with those with SV40 negative tumours.37
Take home messages.
Simian virus 40 (SV40) large T antigen (Tag) DNA sequences were detected in five of 50 historical human hepatocellular carcinomas (HCCs), but were not present in more recent cases of HCC
The presence of Tag sequences may negate the need for p53 mutation and appeared to be associated with a worse prognosis
However, the presence of these sequences is unlikely to be related to exposure to contaminated inactivated polio vaccine
“Regardless of whether simian virus 40 infection can truly promote hepatocarcinogenesis, the presence of the virus or its large T antigen sequence could still theoretically influence phenotype and/or outcome of the malignancy”
If we assume that the presence of SV40 Tag DNA sequences in our patients’ HCCs represents previous infection, a final issue to consider is the source of the virus. The most obvious possibility is exposure to SV40 contaminated inactivated polio vaccine. Although it cannot be assumed that all our patients with HCC were brought up in Scotland, a best estimate of likelihood of exposure to contaminated virus can be gained from past local vaccination policies. Inactivated polio vaccine (later found to be contaminated with SV40) was first used in Scotland in 1956,38 and then replaced in 1961 by oral vaccine,39 from which SV40 has not been isolated.5 Following the policy of primarily vaccinating all children under 15 years of age, individuals most likely to have been exposed to SV40 through this route would therefore have been born between 1941 and 1961. The inactivated vaccine was later also offered to older teenagers and young adults (born after 1933), although take up was poor (< 10%).40 Because none of the five patients with SV40 Tag DNA positive HCC (birth years: 1899, 1905, 1913, 1920, and 1932) was in either age group, they were even less likely to have been inoculated with contaminated vaccine, and so their source of SV40 remains uncertain. However, there is serological evidence of human SV40 infection predating the use of the contaminated inactivated polio vaccine,41 and transmission of SV40 between individuals remains a possibility.4 Interestingly, eight of the 20 cohort B patients were born between 1941 and 1961, yet none of these patients developed HCC with evidence of SV40 Tag DNA.
In conclusion, we have amplified SV40 Tag DNA sequences from a small proportion of historical human HCCs. It is possible that the presence of Tag sequences may substitute for p53 mutation and also associate with a worse prognosis. However, the presence of the sequences is unlikely to be related to exposure to contaminated inactivated polio vaccine and the virus is not present in more recent cases of HCC.
Acknowledgments
We thank Professor J Mullins and Dr S Wharton for their kind gifts of SV40 Tag plasmid and SV40 transgenic mouse neuronal cell line, respectively.
Abbreviations
HCC, human hepatocellular carcinomas
LOH, loss of heterozygosity
PCR, polymerase chain reaction
pRb, retinoblastoma protein
SV40, simian virus 40
Tag, large T antigen
REFERENCES
- 1.London WT. Liver cancer: a disease of the 21st century in America and Europe; lessons from the 20th century in China and Africa [abstract]. In: Abstracts for 6th European meeting on hepatocarcinogenesis: from basic science to treatment, September 23–25 1999, Vienna, Austria. Vienna: University of Vienna, 1999;:I1.
- 2.Anthony PP. Tumours and tumour-like lesions of the liver and biliary tract—aetiology, epidemiology and pathology. In: MacSween RNM, Burt AD, Portmann BC, eds. Pathology of the liver, 4th ed. London: Churchill Livingstone, 2002:711–76.
- 3.Carbone M, Rizzo P, Pass HI. Simian virus 40, poliovaccines and human tumours: a review of recent developments. Oncogene 1997;15:1877–88. [DOI] [PubMed] [Google Scholar]
- 4.Butel JS, Lednicky JA. Cell and molecular biology of simian virus 40: implications for human infections and disease. J Natl Cancer Inst 1999;91:119–34. [DOI] [PubMed] [Google Scholar]
- 5.Sanger DV, Wood DJ, Minor PD. Examination of poliovaccines for the presence of SV40 sequences. In: Brown F, Lewis AM, eds. Simian virus 40 (SV40): a possible human polyomavirus, Dev Biol Stand, Vol. 94. Basel: Karger, 1998:221–5. [PubMed]
- 6.Yamamoto H, Nakayama T, Murakami H, et al. High incidence of SV40-like sequence detection in tumour and peripheral blood cells of Japanese osteosarcoma patients. Br J Cancer 2000;82:1677–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David H, Mendoza S, Konishi T, et al. Simian virus 40 is present in human lymphomas and normal blood. Cancer Lett 2001;162:57–64. [DOI] [PubMed] [Google Scholar]
- 8.Bergsagel DJ, Milton MJ, Butel JS, et al. DNA sequences similar to those of simian virus 40 in ependymomas and choroid plexus tumours of childhood. N Engl J Med 1992;326:988–93. [DOI] [PubMed] [Google Scholar]
- 9.Carbone M, Pass HI, Rizzo P, et al. Simian virus 40-like DNA sequences in human pleural mesothelioma. Oncogene 1994;9:1781–90. [PubMed] [Google Scholar]
- 10.Pacini F, Vivaldi A, Santoro M, et al. Simian virus 40-like DNA sequences in human papillary thyroid carcinomas. Oncogene 1998;16:665–9. [DOI] [PubMed] [Google Scholar]
- 11.Vilchez RA, Madden CR, Kozinetz CA, et al. Association between simian virus 40 and non-Hodgkin lymphoma. Lancet 2002;359:817–23. [DOI] [PubMed] [Google Scholar]
- 12.Shivapurkar N, Harada K, Reddy J, et al. Presence of simian virus 40 DNA sequences in human lymphomas. Lancet 2002;359:851–2. [DOI] [PubMed] [Google Scholar]
- 13.Dubois N, Bennoun M, Allemand I, et al. Time-course of development of well differentiated hepatocarcinoma and lung metastasis in transgenic mice. J Hepatol 1991;13:227–39. [DOI] [PubMed] [Google Scholar]
- 14.Held WA, Pazik J, O’Brien JG, et al. Genetic analysis of liver tumorigenesis in SV40 T antigen transgenic mice implies a role for imprinted genes. Cancer Res 1994:6489–95. [PubMed]
- 15.Casola S, Ungaro P, Pedone PV, et al. Loss of heterozygosity of imprinted genes in SV40 t/T antigen-induced hepatocellular carcinoma. Oncogene 1995;11:711–21. [PubMed] [Google Scholar]
- 16.Lee G-H, Ogawa K, Drinkwater NR. Conditional transformation of mouse liver epithelial cells. An in vitro model for analysis of genetic events in hepatocarcinogenesis. Am J Pathol 1995;147:1811–22. [PMC free article] [PubMed] [Google Scholar]
- 17.Manfredi JJ, Prives C. The transforming activity of simian virus 40 large tumor antigen. Biochim Biophys Acta 1994;1198:65–83. [DOI] [PubMed] [Google Scholar]
- 18.Bannasch P. Pathogenesis of hepatocellular carcinoma: sequential cellular, molecular and metabolic changes. Prog Liver Dis 1996;14:161–97. [PubMed] [Google Scholar]
- 19.Nishida N, Fukuda Y, Ishizaki K, et al. Alteration of cell cycle-related genes in hepatocarcinogenesis. Histol Histopathol 1997;12:1019–25. [PubMed] [Google Scholar]
- 20.Lednicky JA, Butel JS. Consideration of PCR methods for the detection of SV40 in tissue and DNA specimens. In: Brown F, Lewis AM, eds. Simian virus 40 (SV40): a possible human polyomavirus, Dev Biol Stand, Vol. 94. Basel: Karger, 1998:155–64. [PubMed]
- 21.Wright DK, Manos MM. Sample preparation from paraffin-embedded tissues. In: Innis M, Gelfand DH, Sninsky JJ, eds. PCR protocols: a guide to methods and applications. London: Academic Press, 1990:153–8.
- 22.Wong NACS, F Rae, KJ Simpson, et al. Genetic polymorphisms of cytochrome p4502E1 and susceptibility to alcoholic liver disease and hepatocellular carcinoma in a Caucasian population: a study and literature review with meta-analysis. Mol Pathol 2000;53:88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong NACS, Rae F, Bathgate A, et al. Polymorphisms of the gene for microsomal epoxide hydrolase and susceptibility to alcoholic liver disease and hepatocellular carcinoma. Toxicol Lett 2000;115:17–22. [DOI] [PubMed] [Google Scholar]
- 24.Lednicky JA, Stewart AR, Jenkins JJ III, et al. SV40 DNA in human osteosarcomas show sequence variation among T-antigen genes. Int J Cancer 1997;72:791–800. [DOI] [PubMed] [Google Scholar]
- 25.Reuther FJ, Lohler J, Herms J, et al. Low incidence of SV40-like sequences in ependymal tumours. J Pathol 2001;195:580–5. [DOI] [PubMed] [Google Scholar]
- 26.Gurney EG, Tamowski S, Deppert W. Antigenic binding sites of monoclonal antibodies specific to simian virus 40 large T antigen. J Virol 1986;57:1168–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart AR, Lednicky JA, Benzick US, et al. Identification of a variable region at the carboxy terminus of SV40 large T-antigen. Virology 1996;221:355–61. [DOI] [PubMed] [Google Scholar]
- 28.Testa JR, Carbone M, Hirvonen A, et al. A multi-institutional study confirms the presence and expression of simian virus 40 in human mesotheliomas. Cancer Res 1998;58:4505–9. [PubMed] [Google Scholar]
- 29.Sepulveda AR, Finegold MJ, Smith B, et al. Development of a transgenic mouse system for the analysis of stages in liver carcinogenesis using tissue-specific expression of SV40 large T-antigen controlled by regulatory elements of the human alpha-1-antitrypsin gene. Cancer Res 1989;49:6108–17. [PubMed] [Google Scholar]
- 30.Moore M, Teresky AK, Levine AJ, et al. p53 mutations are not selected for in simian virus 40 T-antigen-induced tumors from transgenic mice. J Virol 1992;66:641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall PA, Lane DP. p53 in tumour pathology: can we trust immunohistochemistry? Revisited! J Pathol 1994;172:1–4. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Xu H-J, Murakami Y, et al. Deletions of chromosome 13q, mutations in retinoblastoma 1, and retinoblastoma protein state in human hepatocellular carcinoma. Cancer Res 1994;54:4177–82. [PubMed] [Google Scholar]
- 33.Mayall FG, Jacobson G, Wilkins R. Mutations of p53 gene and SV40 sequences in asbestos-associated and non-asbestos-associated mesotheliomas. J Clin Pathol 1999;52:291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carbone M, Rizzo P, Grimley PM, et al. Simian virus-40 large-T antigen binds p53 in human mesotheliomas. Nat Med 1997;3:908–12. [DOI] [PubMed] [Google Scholar]
- 35.Zhen HN, Zhang X, Bu XY, et al. Expression of the simian virus 40 large tumor antigen (Tag) and formation of Tag–p53 and Tag–pRb complexes in human brain tumors. Cancer 1999;86:2124–32. [PubMed] [Google Scholar]
- 36.Malkin D, Chilton-MacNeill S, Meister LA, et al. Tissue-specific expression of SV40 in tumors associated with the Li-Fraumeni syndrome. Oncogene 2001;20:4441–9. [DOI] [PubMed] [Google Scholar]
- 37.Procopio A, Strizzi L, Vianale G, et al. Simian virus-40 sequences are a negative prognostic cofactor in patients with malignant pleural mesothelioma. Genes Chromosomes Cancer 2000;29:173–9. [DOI] [PubMed] [Google Scholar]
- 38.Report of the Department of Health for Scotland 1958. Edinburgh: Her Majesty’s Stationary Office.
- 39.Ministry of Health. On the state of the Public Health 1962. London: Her Majesty’s Stationary Office.
- 40.Report of the Department of Health for Scotland 1961. Edinburgh: Her Majesty’s Stationary Office.
- 41.Shah K, Nathanson N. Human exposure to SV40: review and comment. Am J Epidemiol 1976;103:1–12. [DOI] [PubMed] [Google Scholar]