Abstract
CO dehydrogenase from the aerobic bacterium Oligotropha carboxidovorans catalyzes the oxidation of CO with H2O, yielding CO2, two electrons, and two H+. Its crystal structure in the air-oxidized form has been determined to 2.2 Å. The active site of the enzyme, which contains molybdenum with three oxygen ligands, molybdopterin-cytosine dinucleotide and S-selanylcysteine, delivers the electrons to an intramolecular electron transport chain composed of two types of [2Fe–2S] clusters and flavin-adenine dinucleotide. CO dehydrogenase is composed of an 88.7-kDa molybdoprotein (L), a 30.2-kDa flavoprotein (M), and a 17.8-kDa iron-sulfur protein (S). It is organized as a dimer of LMS heterotrimers and resembles xanthine dehydrogenase/oxidase in many, but not all, aspects. A mechanism based on a structure with the bound suicide-substrate cyanide is suggested and displays the necessity of S-selanylcysteine for the catalyzed reaction.
CO is a trace gas and important scavenger for hydroxyl radicals in the atmosphere. About one fifth of CO [115–230 Tg⋅yr−1 (1)] is used by soil microorganisms (2, 3). CO has a short lifetime within the soil, and consumption of atmospheric CO occurs mainly in the top few centimeters of the humus layer (O horizon), most likely under aerobic conditions.
Under aerobic conditions, CO can be used as the sole source of carbon and energy by the carboxidotrophic bacteria, which comprise a taxonomically diverse group of obligate or facultative chemolithoautotrophic species (3, 4). Under anaerobic conditions, CO is used with low affinity by a number of methanogenic archaea and acetogenic, sulfate-reducing, or phototrophic bacteria (3, 5). Generally, anaerobic CO dehydrogenases are nickel-containing [4Fe-4S] proteins. CO dehydrogenases of methanogens or acetogens have acetyl-CoA synthetase activity. CO dehydrogenase (EC 1.2.99.2) of the carboxidotrophic bacterium Oligotropha carboxidovorans is a molybdenum-containing iron-sulfur flavoprotein that catalyzes the oxidation of CO (CO + H2O → CO2 + 2e− + 2H+) and generates a proton gradient across the cytoplasmic membrane by channeling the electrons formed via cytochrome b561 into a CO-insensitive respiratory chain (4). O. carboxidovorans uses the reactions of the Calvin cycle for CO2 fixation (4). In the exponential growth phase, cells of O. carboxidovorans have 87% of CO dehydrogenase associated with the inner aspect of the cytoplasmic membrane and 13% located in the cytoplasm (6).
CO dehydrogenase is composed of the 88.7-kDa L (809 residues), 30.2-kDa M (288 residues), and 17.8-kDa S (166 residues) subunits and exists as a dimer of LMS heterotrimers (4). The L subunit carries the molybdenum cofactor, which is a mononuclear complex of Mo and molybdopterin–cytosine dinucleotide (MCD) (7). FAD-binding is on the M subunit and requires conformational changes of M introduced through the binding of M to LS (unpublished data). The S subunit harbors two [2Fe–2S] centers, which are proximal and distal, respectively, to the molybdenum center. They are traditionally designated type I and II according to spectroscopic properties, but the assignment is controversial (8, 9). Recent experiments with M subunit-depleted CO dehydrogenase preparations assign the proximal center to type I and the distal to type II (unpublished data).
CO dehydrogenase has been grouped into the sequence family of molybdenum hydroxylases (10). Other prominent representatives of that family catalyze the oxidative hydroxylation of a diverse range of aldehydes and aromatic heterocycles (9). The high-resolution structure of aldehyde oxidoreductase (Mop) from Desulfovibrio gigas has been solved and has provided an insight into an active site of a molybdenum hydroxylase (11). Mop and CO dehydrogenase both contain the MCD–molybdenum cofactor and the same [2Fe–2S] clusters. However, Mop is devoid of flavin and contains a single polypeptide chain, whereas CO dehydrogenase is a heterotrimeric flavoprotein.
The CO dehydrogenase structural genes (cox) of O. carboxidovorans are clustered in the transcriptional order 5′-coxM-coxS-coxL-3′ along with the Calvin cycle (cbb) and hydrogenase (hox) genes on a 30-kb DNA segment of the 128-kilobase megaplasmid pHCG3 (10, 12). CO dehydrogenase shows significant sequence similarities to the two domains of Mop (10, 11). CoxL is 57% similar (25% identical) to amino acids 175–907 of Mop, and CoxS is 74% similar (41% identical) to amino acids 1–166 of Mop.
MATERIALS AND METHODS
Protein Crystallization.
The protein was crystallized under vapor diffusion conditions by using 0.8 M KH2PO4, 0.8 M NaH2PO4, 2% MPD, and 100 mM Hepes adjusted to pH 7.3. Crystals containing cyanide were cocrystallized in the presence of 4 mM potassium cyanide. For synchrotron data collection under low-temperature conditions, 15% glycerol was added to the crystallization buffer. Before measurements, the concentration of glycerol was gradually increased to 25%. The crystals had an orthorhombic space group P212121 with cell dimensions of a = 129.14 Å, b = 140.34 Å, and c = 164.44 Å and two monomers per asymmetric unit.
Structure Determination.
With a first data set to 3.0 Å, a Patterson search was carried out. The program amore (13) gave a clear solution for a search model constructed from homologous parts of Mop, which comprised a total of about 10% of the scattering mass of the asymmetric unit [R factor of 50.8%, with a correlation coefficient (cc) of 29.4 after translational search for the second monomer, compared with the next highest peak with an R factor of 52.1% and a cc of 24.7 in a resolution range of 14.0–5.0 Å]. Because it was not possible to interpret the calculated density, we initiated multi-wavelength anomalous diffraction data collection at two wavelengths (remote wavelength and iron K-edge wavelength) by using synchrotron radiation at BW6, DESY, Hamburg. Data sets native1 and native2 were collected on a MAR-Research (Hamburg, Germany) charge-coupled device detector at −165°C at BW6, DESY and processed by using denzo and scalepack (14). By using an anomalous difference Fourier map calculated with the phases obtained from the Patterson search solution, the positioning of the four [2Fe–2S] clusters could be achieved. The phasing was carried out by using mlphare (15) and sharp (16) and allowed after solvent flattening and histogram matching the recognition of the molecular boundaries. A phase combination with subsequent solvent flattening and cyclic averaging within the dimer was performed and allowed us to calculate an interpretable electron-density map from the two phase sources. For phase modification, the ccp4 (15) package was used. Density interpretation and model building were carried out by using main (17). The structure was refined by using torsion-angle dynamics, simulated annealing, and maximum likelihood targets for positional refinement as provided by cns (18).
The cyanide-cocrystallized crystal was measured on a MAR-Research (Hamburg, Germany) 300 image plate detector at room temperature by using monochromatized CuK-α radiation produced by a conventional rotating anode (Table 1).
Table 1.
Data collection and refinement statistics
| Native 1/2 | Cyanide | |
|---|---|---|
| Data collection | ||
| Wavelegth, Å | 0.9761/1.7316 | 1.5418 |
| Resolution limit, Å | 2.2/2.5 | 2.36 |
| Unique reflections | 144,247/97,346 | 105,076 |
| Redundancy | 3.4/3.8 | 4.1 |
| Completeness, % | 92.6/89.2 | 92.5 |
| Rsymm* | 0.034/0.053 | 0.119 |
| Refinement | ||
| Protein non-hydrogen atoms | 19,098 | 19,102 |
| Solvent molecules | 1,407 | 783 |
| Resolution range, Å | 25.0–2.2 | 38.0–2.36 |
| Total reflections, F > 2σF | 14,1156 | 10,4855 |
| Total reflections used in Rfree | 4,251 | 3,169 |
| Rcryst,† % | 18.8 | 17.0 |
| Rfrec,‡ % | 23.1 | 20.5 |
| rms deviation of bond distance, Å | 0.0086 | 0.0059 |
| rms deviation of angles, ° | 1.68 | 1.56 |
Rsymm = ∑|I − 〈I〉|/∑I.
Rcryst = ∑∥Fabs| − |Fcalc∥/∑|Fabs|.
Rfree is the same as Rcryst calculated with a test set comprising 5% of the whole data set that was not used in the refinement.
RESULTS
Overall Structure.
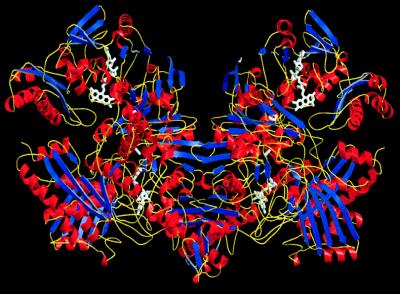
CO dehydrogenase is dimeric in solution, and two monomers were found in the asymmetric unit. The butterfly-shaped dimer (Fig. 1) has overall dimensions of 145 × 100 × 70 Å with an accessible surface area of 78,076 Å2. The two monomers are basically identical, with an rms deviation of 0.30 Å for the Cα atoms.
Figure 1.
Ribbon representation of the CO dehydrogenase dimer. The twofold-symmetry axis of the dimer runs vertically in the plane of the figure. Helices are shown in red, β-sheets in blue, and the connecting turns and loops in yellow. The cofactors are depicted in white. Structure presentations were created by using bobscript and raster3d.
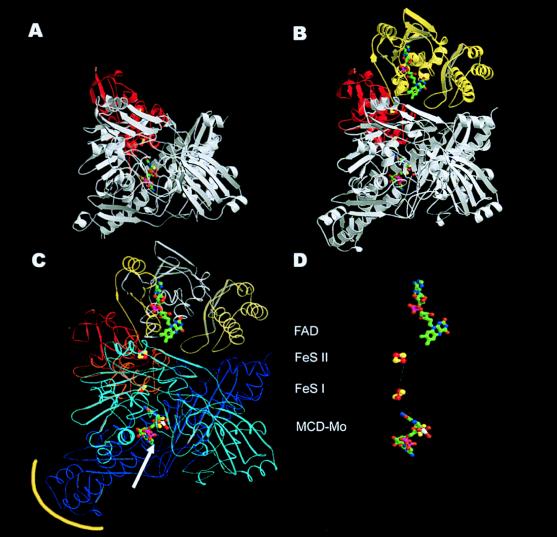
The redox components of one LMS-structured monomer are the MCD–molybdenum cofactor, composed of a molybdenum ion with two oxo- and one hydroxoligand, complexed by the enedithiolene group of MCD, [2Fe–2S] clusters of type I and type II, and a noncovalently bound FAD molecule (Fig. 2 C and D). In air-oxidized CO dehydrogenase, the flavin is fully oxidized, the oxidation state of Mo is +VI, and MCD occurs in a redox state that is reduced by two electrons compared with the fully oxidized state (19), a tricyclic tetrahydropterin–pyran system. The MCD–molybdenum cofactor is located in the L subunit, and the two [2Fe–2S] clusters are located in the S subunit (Fig. 2). These prosthetic groups form a pathway for the electrons to the FAD located in the M subunit (Fig. 2). The two active sites of the dimer are at a Mo–Mo distance of 52.9 Å with no further cofactors located between them, indicating two separate catalytic units in CO dehydrogenase.
Figure 2.
Comparison of the CO dehydrogenase monomer (B) with the monomeric Mop structure from Desulfovibrio gigas (12) (A). Domain structure (C) and cofactors (D) of the CO dehydrogenase monomer. The ribbon presentation shows the general similarity of the two monomers for the MCD–molybdenum cofactor-containing subunit/domain (white) and the iron-sulfur containing subunit/domain (red) in both enzymes. The FAD-containing M subunit (yellow) is absent in the Mop protein. Whereas the S subunits are very similar in both proteins, the L subunit differs in a few areas. This applies mainly to the substrate channel of Mop (white arrow), which is closed by an additional loop in CO dehydrogenase and to the first part of the C-terminal domain (yellow encircled), which creates the dimer interface. (C) The three subunits of CO dehydrogenase may be subdivided into domains (the L subunit in a cyan N-terminal domain and a blue C-terminal domain). The S subunit has the iron-sulfur clusters bond in the N-terminal domain drawn in red and in the C-terminal domain drawn in orange. The three domains of the M subunit are yellow, white, and cream, sequentially, from N to C terminus. (D) The shortest connection between the cofactors is marked by a white line, starting from the molybdo-oxo group (MCD–Mo) at the active site via the proximal C-terminal [2Fe–2S] cluster (FeS I) (14.6 Å distance from the Mo atom to the closest iron atom) and the distal N-terminal [2Fe–2S] cluster (FeS II) (12.4 Å distance between the closest iron atoms), ending at the FAD (8.7 Å distance between C7 of FAD and the closest iron atom).
Structure of the Three Subunits.
Subunit S represents the iron-sulfur protein of CO dehydrogenase and is clearly divided into a C- and an N-terminal domain, each binding a [2Fe–2S] cluster, which can be distinguished by EPR (8). The C-terminal domain (residues 77–161) carries the proximal [2Fe–2S] cluster. The cluster is buried in CO dehydrogenase ≈11 Å below the protein surface at the interface between the S and the L subunit and is adjacent to the MCD–molybdenum cofactor. The [2Fe–2S] cluster is located at the N terminus of two α-helices that participate in a four-helix bundle of twofold symmetry, an architecture first described for the Mop protein (11).
The N-terminal domain (residues 3–76) is similar to those of plant-type [2Fe–2S]-ferredoxins (20). It carries the distal [2Fe–2S] cluster, which is adjacent to the FAD residing on M. Although the cluster is positioned at the interface between S and M, it is exposed to the solvent and mediates electron transfer from the proximal [2Fe–2S] cluster to the isoalloxazine ring of FAD.
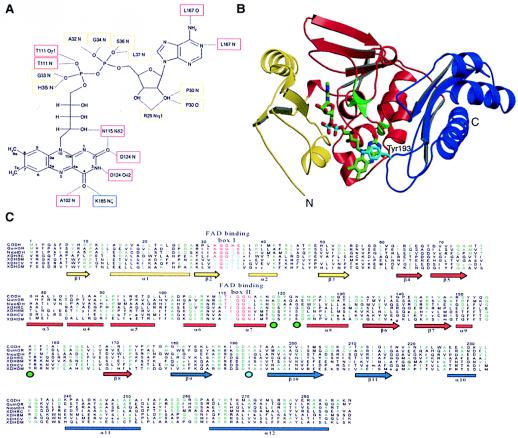
Subunit M is the flavoprotein of CO dehydrogenase. It binds FAD in a 1:1 molar stoichiometry (Fig. 5 A and B) and exhibits the motifs 32MAGGHS36M and 111MTIGG114M typical of the FAD-binding site of the vanillyl-alcohol oxidase family of flavoproteins. Both motifs are conserved in the molybdenum hydroxylase sequence family (Fig. 5C). M is arranged in three distinct domains (Fig. 5 B and C), designated the N-terminal domain (residues 1–54), the middle domain (residues 60–174), and the C-terminal domain (residues 180–285). The N-terminal domain is composed of a three-stranded parallel β-sheet (β1–β3) flanked by two helices (α1–α2). A loop between α2 and β2 of a βαβ-unit (P loop, residues 32–36) contains one of the two double-glycine motifs, which interact with the pyrophosphate moiety of the FAD molecule. The main structural element of the middle domain is a five-stranded antiparallel β-sheet (β4–β8) that contains an insertion of six small α-helices winding around the adenosine dinucleotide portion of the FAD molecule. A small α-helix contains the second double-glycine motif (residues 111–115), which also interacts with the FAD. The N-terminal and the middle domains form the wings holding the cofactor (Fig. 5B).
Figure 5.
Structure of the M subunit. (A) Molecular interactions of FAD in the M subunit. Atoms of the isoalloxazine ring are labeled. Residues binding the FAD cofactor through hydrogen bonds are marked, and boxes around the labels give the colors of the domains that harbor the corresponding residue. Whereas the N-terminal domain (yellow boxes) binds only to the dinucleotide portion of FAD, the middle domain (red boxes) interacts also with the riboflavin part. The C-terminal domain (blue box) shows only a single hydrogen bond to FAD. (B) Fold of the M subunit. The N-terminal domain is shown in yellow, the middle domain in red, and the C-terminal domain in blue. Tyr193M shields the isoalloxazine ring and is shown in cyan. The positions of residues corresponding to the rosy mutant strains of Drosophila melanogaster that resulted in an altered affinity to bind NAD+ in xanthine dehydrogenase are shown as green enlargements of the Cα chain and have a green circle under their position in C. (C) Sequence alignment of the M polypeptide of CO dehydrogenase with the corresponding representative parts from other members of the molybdenum hydroxylase family: CODH, CO dehydrogenase from Oligotropha carboxidovorans (coxM, Swall: Q51323); QuinOR, quinoline 2-oxidoreductase from Pseudomonas putida (qorM, Swall: P72222); NicotDH, nicotine dehydrogenase from Arthrobacter nicotinovorans (ndhA, Swall: Q59127); XDHRC, xanthine dehydrogenase from Rhodobacter capsulatus (xdhA, Swall: O54050); XDHBM, xanthine dehydrogenase from Bombyx mori (Swall: Q17209); XDHCV, xanthine dehydrogenase from Calliphora vicina (xdh, Swall: XDH CALVI); XDHDM, xanthine dehydrogenase from Drosophila melanogaster (xdh, Swall: XDH DROME). Two boxes highlighting residues responsible for FAD binding in the M subunit are shown in red and blue. They correspond to fingerprints for FAD-binding by enzymes of the molybdenum hydroxylase family and define a FAD-binding motif similar to the motifs found for the vanillyl alcohol oxidase family of flavoproteins. The secondary structure is represented by arrows for β-sheets and bars for α-helices, in domain colors (B). Residues appearing at least three times in a row are colored in green. The position of Tyr193M is marked by a cyan circle below the aligned sequences.
The C-terminal domain contains a three-stranded antiparallel β-sheet (β10–β12) that merges in a bundle of three α-helices (α8–α10). It is obvious that the isoalloxazine ring is accessible only from one side of the M subunit near the subunit interface. The central part of the isoalloxazine ring is shielded from the solvent by the side chain of Tyr193M, which is part of an extended loop created between β10 and β11 (Q loop, residues 192–195) of the C-terminal domain (Fig. 5 B and C). The distal parts of the dimethyl benzene ring remain solvent accessible. This applies to the C7 and C8 positions with their methyl groups as well as for the C9 position of the isoalloxazine ring. The C4a and N5 atoms may become accessible by movements of the C-terminal domain or the side chain of Tyr193M. The C-terminal domain is linked to the middle domain by a flexible loop that could provide the flexibility. The C-terminal domain interacts only with the isoalloxazine ring at its O4 position.
By using the program dali (21), two proteins were identified with similarity to the M subunit of CO dehydrogenase with respect to the architecture of structural elements interacting with FAD. MurB (UDP–N-acetylenolpyruvylglucosamine reductase) of Escherichia coli (22) showed a Z score of 8.9 by using the fold of M as a search model. The vanillyl–alcohol oxidase (VAO) of Penicillium simplicissimum (23) gave a Z score of 6.0. VAO is a member of a new oxidoreductase family sharing a conserved FAD-binding domain (24). The fold of MurB and VAO involved in binding the dinucleotide moiety of the FAD cofactor resembles the N-terminal and middle domain of the M subunit of CO dehydrogenase. The residues forming two loops responsible for binding the FAD in the CoxM polypeptide are mainly conserved in the molybdenum hydroxylase enzyme family (Fig. 5C) and can also be found as conserved fingerprints in the new VAO family. Both loops have in their central portion two small amino acids, often a double glycine. The first binding motif lying on the P loop can be defined as aGgt, with the sequence 32AGGH35 in CoxM. The second motif on α6 and α7 can be defined as AxpqiRnxatxGGn, equivalent to 102ADPQIRYMGTIGGN115 in CoxM (Fig. 5C). In the VAO family, a ssGHs and a shsG motif are responsible for binding the pyrophosphate moiety of FAD (24). It is likely that other flavin-containing molybdenum hydroxylases exhibit flavin-binding sites similar to that observed in CO dehydrogenase.
The C-terminal domains of the CO dehydrogenase M subunit and its counterparts in MurB and VAO are structurally unrelated and may convey class-specific properties.
Subunit L has a heart-like shape with dimensions of 60 × 83 × 70 Å and contains two subdomains (Fig. 2C). Its N-terminal domain (residues 10–439) interacts with the M and S subunits in the same monomer. Its fold is dominated by a five- and a four-stranded mixed β-sheet. The C-terminal domain (residues 440–809) can be further subdivided into two parts. The first part consists of a central α-helix of 30 aa that runs into a small three-stranded β-sheet followed by an α-helix and a four-stranded parallel–antiparallel β-sheet, which interacts with its symmetrymate of the other monomer, providing the main dimer contact. The second part of the C-terminal domain consists of a long three-stranded antiparallel β-sheet surrounded by seven α-helices.
Active Site with S-Selanylcysteine.
The active site is accessible from the outside through a narrow channel that is 17 Å deep with an average diameter of 7 Å, determined from the Cβ of the surrounding residues. A restriction of the active-site channel may exclude substrates larger than CO and may prevent dissociation of the cofactor from the protein. The character of the channel is hydrophobic, so that the first charged residues an incoming molecule can interact with are the protein ligands around the molybdenum in its second coordination sphere.
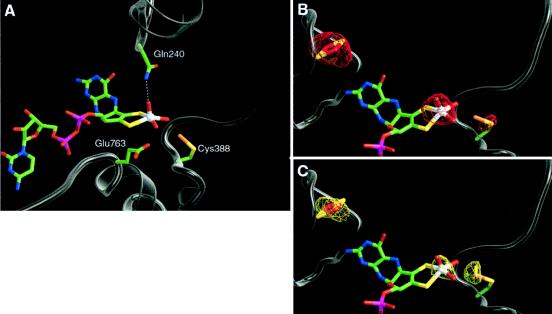
The MCD–molybdenum cofactor is buried at the center of the L subunit (Fig. 2C) and is ligated through a dense network of hydrogen bonds originating from both domains of L. The geometry of the first coordination sphere around the Mo ion can be described as a distorted square pyramid (Fig. 3A). The dithiolene group of MCD is positioned in the equatorial plane together with two oxygen atoms, which were modeled as an oxo- and a hydroxo- group. The apical ligand was modeled as an oxo- group. In contrast to xanthine oxidases/dehydrogenases, active CO dehydrogenase does not contain a cyanolysable sulfido ligand at the Mo ion.
Figure 3.
The molybdenum site of the L subunit. (A) The Mo ion exhibits one hydroxo and two oxo ligands and is bound to MCD through its enedithiolene group. The second coordination sphere of the Mo ion is composed of the residues Gln240L, Glu763L, and S-selanyl-Cys388L. B and C show the active site together with the adjacent [2Fe–2S] cluster. Difference Fourier maps were calculated with the phases from the final model shifted by 90° (imaginary map) and with amplitudes from the Bijvoet differences at a wavelength λ1 = 1,7316 Å (B) shown in red and calculated with data obtained at a wavelength of λ2 = 0,9761 Å (C) shown in yellow. The maps are contoured at 3σ. Calculated f′′ values using the program absorb for selected free atoms are Fe: λ1, 3.9 e− and λ2, 1.5 e−; Se: λ1, 1.41 e− and λ2, 3.84 e−; S: λ1, 0.69 e− and λ2, 0.23 e−; Mo: λ1, 3.36 e− and λ2, 1.23 e−.
The apical oxo-group can form a hydrogen-bond to the Nɛ2 of Gln240L, located in a distance of 3.0 Å. Gln240L is highly conserved in the molybdenum hydroxylase family, although it has no equivalent in the Mop protein.
Trans to the apical oxo group is a conserved glutamate residue (Glu763L) with a Mo–Oɛ1 distance of 3.1 Å. The second coordination sphere contains a modified cysteine residue (Cys388L, Fig. 3 A–C). Strong additional density between Cys388L and the equatorial oxo group was modeled as a SeH group attached to the Sγ of the cysteine residue as an S-selanylcysteine. The identification of the selenium is based on the anomalous scattering at two different wavelengths (λ1 = 0.976 Å and λ2 = 1.732 Å, Fig. 3 B and C) and agrees with data of chemical analyses (ref. 25; unpublished results). The selenium atom of S-selanylcysteine is located in a distance of 3.7 Å from the Mo ion. It is near the equatorial oxo and hydroxo group of the Mo ion. The modified residue is situated at a loop that shows an amino acid composition unique for the CO dehydrogenases, because its sequence of VAYRC388LSFR is also present in the sequence of CO dehydrogenase from Pseudomonas thermocarboxydovorans or Hydrogenophaga pseudoflava but absent in other enzymes of the molybdenum hydroxylase family. The active-site loop occupies the position of the substrate-analog isopropanol bound in the Mop structure and could be involved in substrate-binding in CO dehydrogenase.
DISCUSSION
Hypothetical Reaction Mechanism.
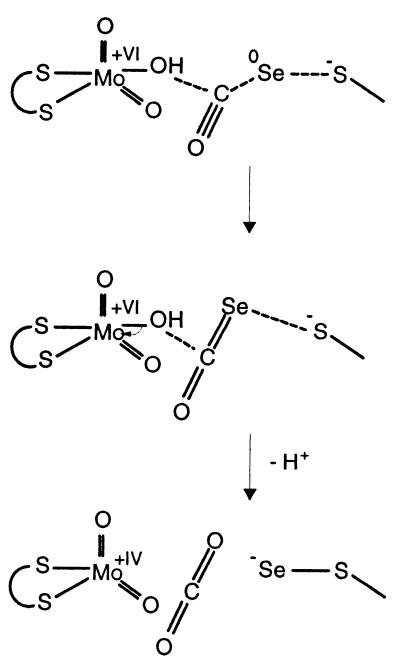
The chemistry of carbon oxide selenide (O⩵C⩵Se) and the selenium-catalyzed synthesis of carbonates from CO (26) show remarkable analogies to the CO dehydrogenase-catalyzed oxidation of CO to CO2.
SeCO is a gas, which is easily prepared through the oxidation of CO with elemental selenium in the presence of a strong base at normal pressure and room temperature (26). When SeCO is treated with a sodium alcoholate (RONa) under mild conditions, the corresponding sodium O-alkyl selenocarbonate (ROC⩵OSe−Na+) is formed, which undergoes nucleophilic attack by the alcohol, yielding the corresponding dialkyl carbonate [(RO)2C⩵O]. In aqueous sodium hydroxide (r = H), carbon oxide selenide reacts to carbonate and selenide (COSe + 4 NaOH → Na2Se + Na2CO3 + 2 H2O).
The active site of CO dehydrogenase literally assembles all components essential in the selenium-catalyzed synthesis of carbonates (Fig. 3). The selenium of the S-selanylcysteine388L is formally elemental Se with the redox state 0 (Fig. 4). It reacts with cyanide, yielding selenocyanate, with the concomitant irreversible inactivation of CO dehydrogenase under oxic or anoxic conditions (half-life of 1 h, complete inactivation after 10 h). Enzyme crystallized in the presence of 4 mM potassium cyanide showed a reduced occupancy, with selenium at the S-selanylcysteine residue in the structure, and the remaining selenium now appears in a small linear molecule interpreted as selenocyanate. The molecule lies in the equatorial plane of the oxygen ligands, and the selenium is in hydrogen-bond distance to the amide nitrogen atoms of Cys388L and Ala385L. Because CO and cyanide are isoelectronic, isosteric, and show a similar chemical reactivity, we assume a similar reaction between CO and selenium, leading to a carbonyl selenide that reacts with water to yield CO2 (Fig. 4).
Figure 4.
Hypothetical reaction mechanism. Product CO2 may remain in the metal coordination sphere such that the number of oxygen ligands is unchanged during the catalytic steps.
The catalytic mechanism of CO dehydrogenase thus can be imagined to involve the formation of carbon oxide selenide with the subsequent nucleophilic attack by the Mo–OH group, yielding CO2 (Fig. 4). Catalysis in CO dehydrogenase starts by the reaction of CO with the selenium of S-selanylcysteine388L, thereby forming a selenocarbonyl species (Fig. 4). The selenocarbonyl undergoes a nucleophilic attack by the Mo–OH group of the molybdenum, which is in the oxidized +VI state. Subsequently, two electrons are abstracted, and CO2 is released. Electron transfer from the reduced Mo ion in the +IV state to the proximal [2Fe–2S] center and the addition of water regenerates the oxidized Mo–OH in the +VI oxidation state.
The transport of the electrons from the active-site Mo ion to the external electron acceptors is facilitated through a chain of redox active cofactors composed of two [2Fe–2S] clusters and FAD. The MCD–molybdenum cofactor is adjacent to the proximal [2Fe–2S] cluster located in the C-terminal domain of the S subunit (Fig. 2D). The molybdopterin ring is positioned between the molybdenum and the [2Fe–2S] cluster and supports electron transfer through its conjugated double bonds. The N2′ amino group of the fused pyrimidin ring is at a distance of 5.5 Å from the Fe1 atom of the proximal cluster.
CO Dehydrogenase as a Member of the Xanthine Oxidase Family.
The CO dehydrogenase reveals significant sequence homologies with the other members of the molybdenum hydroxylase family, suggesting structural conservation and represents, therefore, a close structural model of the eukaryotic xanthine oxidase/dehydrogenase in tertiary and quarternary structure. Mop had served as a first approximation to the structure of the xanthine oxidase/dehydrogenase (11), but lacked the flavin domain that is now defined in CO dehydrogenase by the M subunit. Its location differs from the position of the connecting loop in Mop suggested to be a marker (11). Major gaps in sequence alignments between the enzymes are due to the fact that most of the xanthine oxidases/dehydrogenases, as well as Mop, contain connecting segments between their structural domains, which are missing in the heterotrimeric CO dehydrogenase.
The homology between CO dehydrogenase and other members of the molybdenum hydroxylase family extends beyond to residues in the core of the domains/subunits to the interdomain/subunit contacts. In addition, the arrangement of the L and the S subunit of CO dehydrogenase is identical to the arrangement of the corresponding parts of Mop (Fig. 2).
The major functional difference between the interconvertible forms xanthine oxidase and xanthine dehydrogenase is their usage of electron acceptors. These different reactivities may be correlated to the ability of binding NAD+/NADH. Affinity-labeling studies with the pyridine nucleotide analog 5′-p-fluorosulfonylbenzoyladenosine (5′-FSBA) (27) hinted to a tyrosine residue affecting NAD+ binding. Reactivity studies of xanthine dehydrogenase variants from rosy mutant strains of Drosophila melanogaster comprised mutants that showed a diminished affinity for NAD+ or were unable to use NAD+ as an electron acceptor (28). Through sequence comparisons between the xanthine oxidase/dehydrogenase sequences from different sources with the sequence of the CO dehydrogenase M subunit, we localized in our model the positions of these mutations and the labeled tyrosine residue. They correspond to Gly119M, Asn123M, and Ala156M in CoxM (Fig. 5 B and C). All of these residues cluster around two loops of the M subunit near the solvent-exposed side of the isoalloxazine ring of the FAD. Their mutation may alter the ability to bind NAD+. The likely docking site of NAD+ suggests hydride transfer from the isoalloxazine ring to a pyridine ring positioned at the si face. This unusual stereochemistry was also suggested for the interaction of MurB with NADP+ (29).
Acknowledgments
We thank Dr. H. Bartunik and G. B. Bourenkow at the BW6, DESY, Hamburg for their help in data collection and multiwavelength anomalous diffraction measurements. This work was supported by Grant Me 732/5-3 from the Deutsche Forschungsgemeinschaft (Bonn-Bad Godesberg, Germany) to O.M.
ABBREVIATIONS
- MCD
molybdopterin–cytosine dinucleotide
- VAO
vanillyl–alcohol oxidase
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1QJ2).
References
- 1.Sanhueza E, Dong Y, Scharffe D, Lobert J M, Crutzen P J. Tellus, Ser B. 1998;50:51–58. [Google Scholar]
- 2.Moxley J M, Smith K A. Soil Biol Biochem. 1998;30:65–79. [Google Scholar]
- 3.Mörsdorf G, Frunzke K, Gadkari D, Meyer O. Biodegradation. 1992;3:61–82. [Google Scholar]
- 4.Meyer O, Frunzke K, Mörsdorf G. In: Microbial Growth on C1 Compounds. Murrell J C, Kelly D P, editors. Andover, U.K.: Intercept; 1993. pp. 433–459. [Google Scholar]
- 5.Ragsdale S W, Kumar M. Chem Rev. 1996;96:2515–2539. doi: 10.1021/cr950058+. [DOI] [PubMed] [Google Scholar]
- 6.Rohde M, Mayer F, Meyer O. J Biol Chem. 1984;259:14788–14792. [PubMed] [Google Scholar]
- 7.Meyer O, Frunzke K, Tachil J, Volk M. In: Molybdenum Enzymes, Cofactors, and Model Systems. Stiefel E I, Coucouvanis D, Newton D W E, editors. Washington, DC: Am. Chem. Soc.; 1993. pp. 433–459. [Google Scholar]
- 8.Bray R C, George G N, Lange R, Meyer O. Biochem J. 1983;211:687–694. doi: 10.1042/bj2110687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hille R. Chem Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 10.Schübel U, Kraut M, Mörsdorf G. J Bacteriol. 1995;177:2197–2203. doi: 10.1128/jb.177.8.2197-2203.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romao M J, Archer M, Moura I, Mowra J J G, LeGall J, Engh R, Schneider M, Hof P, Huber R. Science. 1995;270:1170–1176. doi: 10.1126/science.270.5239.1170. [DOI] [PubMed] [Google Scholar]
- 12.Santiago B, Meyer O. J Bacteriol. 1997;179:6053–6060. doi: 10.1128/jb.179.19.6053-6060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navazza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 14.Otwinowski Z. In: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S, editors. Daresbury Laboratory, Warrington, U.K.: Science and Engineering Research Council; 1993. pp. 56–62. [Google Scholar]
- 15.Collaborative Computational Project No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 16.de la Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–493. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 17.Turk D. Ph.D. thesis. Germany: Technische Universität München; 1992. [Google Scholar]
- 18.Bruenger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.Tachil J, Meyer O. FEMS Microbiol Lett. 1997;148:203–208. [Google Scholar]
- 20.Sticht H, Roesch P. Prog Biophys Mol Biol. 1998;70:95–136. doi: 10.1016/s0079-6107(98)00027-3. [DOI] [PubMed] [Google Scholar]
- 21.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 22.Benson T E, Filman D J, Walsh C T, Hogle J M. Nat Struct Biol. 1995;2:644–653. doi: 10.1038/nsb0895-644. [DOI] [PubMed] [Google Scholar]
- 23.Mattevi A, Fraaije M W, Mozzarelli A, Olivi L, Coda A, van Bokel W J H. Structure (London) 1997;5:907–920. doi: 10.1016/s0969-2126(97)00245-1. [DOI] [PubMed] [Google Scholar]
- 24.Fraaije M W, Berkel W J H, van Benen J A E, Visser J, Mattevi A. Trends Biochem Sci. 1998;23:206–207. doi: 10.1016/s0968-0004(98)01210-9. [DOI] [PubMed] [Google Scholar]
- 25.Meyer O, Rajagopalan K V. J Biol Chem. 1984;259:5612–5617. [PubMed] [Google Scholar]
- 26.Paulmier C. Selenium Reagents and Intermediation in Organic Synthesis. Oxford: Pergamon; 1986. [Google Scholar]
- 27.Nishino T, Nishino T. Biochemistry. 1987;26:3068–3072. doi: 10.1021/bi00385a018. [DOI] [PubMed] [Google Scholar]
- 28.Doyle W A, Burke J F, Chovnick A, Dutton F L, Whittle J R S, Bray R C. Eur J Biochem. 1996;239:782–795. doi: 10.1111/j.1432-1033.1996.0782u.x. [DOI] [PubMed] [Google Scholar]
- 29.Constantine K L, Mueller L, Goldfarb V, Wittekind M, Metzler W J, Yanchunas J, Jr, Robertson J G, Malley M F, Fridericks M S, Farmer B T, II. J Mol Biol. 1997;267:1223–1246. doi: 10.1006/jmbi.1997.0915. [DOI] [PubMed] [Google Scholar]