Abstract
Aims: To develop an alternative assay for specific genotyping of the −α4.2 thalassaemia deletion based on the DNA sequence features surrounding the breakpoint.
Methods: The 5′ and 3′ ends of the breakpoint regions of the −α4.2 allele and the normal homologous segments were sequenced in Chinese individuals. A sequence haplotype composed of four single nucleotide variations within the X2/X1 box of the −α4.2 breakpoint region was found in all of the 10 Chinese −α4.2 thalassaemia alleles studied. Based on these findings, a novel polymerase chain reaction (PCR)/denaturing high performance liquid chromatography (DHPLC) assay was developed for rapid genotyping of the −α4.2 allele instead of traditional Southern blotting or Gap-PCR. This method involves amplification of the α globin target sequence encompassing these four polymorphic sites, followed by a partially denaturing HPLC analysis using the transgenomic WAVE DNA fragment analysis system.
Results: The three major genotypes (−α4.2/αα, −α4.2/--SEA, and αα/αα) could be distinguished through the characteristic chromatograms generated by the WAVE system. The accuracy of this technique was evaluated blindly, and the results were 100% (40 of 40) concordant with the genotypes previously characterised by Southern blotting or Gap-PCR.
Conclusions: This study validates the PCR/DHPLC approach as a simple, rapid, highly accurate, and cost effective method, potentially adaptable for use in epidemiological surveys, genetic screening, and diagnosis of silent α+ thalassaemia and Hb H disease.
The human α globin gene cluster on chromosome 16 consists of seven gene members including two major α globin genes—α2 and α1—which have almost identical coding sequences and are embedded within two highly homologous 4 kb duplicated units. These regions are divided into three homologous subsegments—X, Y, and Z boxes—by the non-homologous elements (I, II, and III).1,2Crossovers between misaligned X or Z boxes give rise to −α4.2 or −α3.7 chromosomes. These two deletional alleles are the most common single gene determinants producing asymptomatic/silent carriers of α thalassaemia. In particular, α+ thalassaemia, resulting from the −α4.2 deletion, is mainly distributed in certain areas of the tropics and subtropics including Melanesia, India, Southeast Asia, and southern China, with estimates of gene frequencies ranging from 2% to 60%.3–5
“In our study, we aimed to develop a new assay for the specific identification of the −α4.2 deletion based on the analysis of DNA sequence features around the breakpoint”
In Southeast Asia and southern China, the incidence of Hb H disease, which is caused by the co-inheritance of the cis α0 thalassaemia deletion and the α+ thalassaemia determinant, is relatively frequent because both the α0 thalassaemia deletion (--SEA/) and α+ thalassaemia alleles (−α4.2/ and −α3.7/) are very common in the population.6 Traditionally, Southern blotting is the standard method used to detect gene deletions. However, it is time consuming, labour intensive, and is not feasible for large scale population screening.7 In recent years, several techniques for the molecular characterisation of common α thalassaemia deletions have been described.8–11 The Gap-polymerase chain reaction (PCR) technique, based on the amplification of the junction segment of the breakpoint, is a notable example. However, there are problems with the reproducibility of this technique because PCR does not perform well for relatively long fragments with high G/C content. Thus, there is room for the development of new techniques that are highly accurate, rapid, inexpensive, automated, and easily interpretable, to aid in the diagnosis of α thalassaemia traits. This is especially relevant to the genotyping of α+ thalassaemia deletions for epidemiological surveys, genetic screening, and the diagnosis of silent α thalassaemia carriers and Hb H disease, where there are no detectable phenotypic features. At present, the diagnosis of α+ thalassaemia is only possible through the detection of Hb Bart’s in cord blood samples by electrophoretic analysis.12
The analysis of DNA by denaturing high performance liquid chromatography (DHPLC) is a relatively new method for the rapid detection and identification of point mutations.13,14In our study, we aimed to develop a new assay for the specific identification of the −α4.2 deletion based on the analysis of DNA sequence features around the breakpoint. Hence, we sequenced the breakpoint region of the −α4.2 allele and the normal homologous segments encompassing both the 5′ and 3′ ends in Chinese individuals. It is interesting that four single nucleotide variations were found to be linked to the −α4.2 haplotype. Based on these new findings, a novel DHPLC assay for the rapid detection/genotyping of the −α4.2 deletion by the profiling of short fragment amplicons was developed. The feasibility of this assay for the detection of −α4.2 alleles was tested in 40 samples with various genotypes involving the −α4.2 allele and the results were compared with that of Gap-PCR or DNA sequence analysis.
MATERIALS AND METHODS
Subjects
The samples were collected from different geographical regions in southern China, namely the Guangdong Province and Guangxi Zhuang Autonomous Region, where the thalassaemias are highly prevalent. Genomic DNA was extracted from peripheral blood leucocytes by standard procedures. The sequences around the breakpoint regions and the normal homologous segments were determined in 14 DNA samples with known −α4.2/αα (eight cases), −α4.2/--SEA (two cases), or αα/αα (four cases) genotypes. Six of the cases with three different known genotypes (two each of −α4.2/αα, −α4.2/--SEA, and αα/αα) were used for the methodological development of the novel DHPLC assay. The genotypes of each of these six cases were characterised by Southern blotting analysis. Subsequently, a total of 40 blood and prenatal archival samples with −α4.2 alleles or normal α globin gene sequences, which had previously been identified by Gap-PCR or Southern blotting, were used to test the performance of the our new assay by blind analysis. These 40 samples encompassed many different genotypes, including 16 heterozygous carriers, 10 patients with Hb H disease caused by co-inheritance of the cis α0 thalassaemia deletion and α+ thalassaemia allele (--SEA/−α4.2), and 14 normal individuals.
DNA sequence analysis
The primers for amplifying the breakpoint region of the −α4.2 allele were P71 (5′-TACCCATGTGGTGCCTCCATG-3′) and P72 (5′-TGTCTGCCACCCTCTTCTGAC-3′), previously described by Oron-Karni et al,8 which produced a 1596 bp PCR amplified product. The primers for amplifying the normal homologous segment in X box 2 were P71 and P3 (5′-CCGACCTCAGGTGATCC TCT-3′), which produced a 1302 bp PCR amplified product. P5 (5′-GTTAAGCTGGA GCCTCGGTAG-3′) and P72 were used to amplify the normal homologous segment in X box 1, producing a 1519 bp fragment. P3 and P5 were designed based on the α globin cluster sequence (accession number NG_000006) from National Center for Biotechnology Information (NCBI) GeneBank. Figure 1 shows the locations of these primers.
Figure 1.
Locations of the polymerase chain reaction primers used for DNA analysis (P71, P3, P5, and P72) and denaturing high performance liquid chromatography (D1 and D2) in (A) the normal α globin gene cluster and (B) the −α4.2 chromosome produced by unequal crossing over between the two X boxes.
PCR was performed in a total volume of 50 μl containing 20 mmol/litre Tris/HCl (pH 8.4), 50 mmol/litre KCl, 1.5 mmol/litre MgCl2, 0.2 μM of each primer, 0.2 mmol/litre of each dNTP, 2.0 U of DNA polymerase (TaKaRa, LA Taq™; TaKaRa Biotechnology, Dalian, China), and 100 ng of genomic DNA. Reactions were carried out in a thermal cycler (PCR express thermal cycler; Hybaid Ltd, Ashford, Middlesex, UK), with an initial three minutes of denaturation at 95°C; 30 cycles of 95°C for 30 seconds, 62°C for 40 seconds, 72°C for one minute and 30 seconds; and a final extension at 72°C for three minutes.
PCR products amplified from the 14 samples (10 samples with −α4.2 alleles and four αα haplotypes) used for the breakpoint sequence verification part of the study were directly sequenced using an ABI 377 sequencer (PE Biosystems; Foster City, California, USA) with BigDye terminator kit (PE Biosystems). The DNA sequences of the −α4.2 breakpoint region were aligned with the homologous sequences of the X boxes from both the Chinese healthy individuals we sequenced and the Homo sapiens genomic α globin region (HBA@) on chromosome 16 (NG_000006, GeneBank, NCBI) using the software DNAMAN 4.0 (Lynnon BioSoft Inc, Vandreuil, Quebec, Canada). The homologous DNA sequence from NG_000006 is from nt 30310 to nt 31656 (X2 box) or from nt 34567 to nt 35911 (X1 box).15,16
PCR conditions for DHPLC analysis
The primers used to produce the amplicons for DHPLC analysis were designed according to the newly found nucleotide variations linked to the −α4.2 allele in our study. These four variations were within the X2/X1 box of the −α4.2 breakpoint region. We designed a PCR system to amplify the DNA segment encompassing these four polymorphisms. The primers were D1 (5′-CTCTGGGACCTCC TGGTGCTT-3′) and D2 (5′-CAGGAAGAGCGGGTGGTGGAG-3′) (fig 1), which produced a 312 bp PCR amplified fragment. Each 50 μl PCR mix contained the following components: 20 mmol/litre Tris/HCl, pH 8.4, 50 mmol/litre KCl, 1.5 mmol/litre MgCl2, 25 pmol of each primer, 0.2 mmol/litre of each dNTP, 2 U polymerase (TaKaRa, γ q™; TaKaRa Biotechnology), and 100 ng of genomic DNA. The reaction was carried out on a thermal cycler (PCR Express; Hybaid) with an initial three minutes of denaturation at 95°C; 30 cycles of 95°C for 30 seconds, 62°C for 40 seconds, and 72°C for 30 seconds; and a final extension at 72°C for three minutes.
DHPLC analysis
DHPLC analysis was conducted on the automated WAVE™ nucleic acid fragment analysis system (Transgenomic, Omaha, Nebraska, USA). The PCR product (5 μl) was injected into a preheated (61°C) reversed phase column based on non-porous poly (styrene/divinylbenzene) particles (DNASep column; Transgenomic), and eluted from the column by a linear acetonitrile gradient at a flow rate of 0.9 ml/minute for seven minutes. The linear acetonitrile gradient consisted of 45% buffer A (0.1 mmol/litre triethyl-ammonium acetate (TEAA); pH 7.0) and 55% buffer B (0.1 mol/litre TEAA; pH 7.0, containing 25% acetonitrile), and buffer B increased at 2%/minute. Fragments were eluted with the temperatures that were calculated by the DHPLC melt program for the successful resolution of heteroduplexes (http:/insertion.stanford.edu/). The DNA samples were typed by analysis of DHPLC distinct profiles.
RESULTS
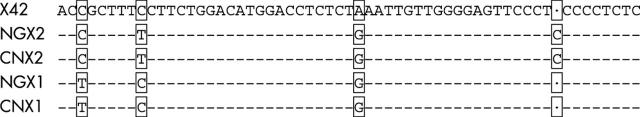
Sequence alignment analysis showed that the DNA sequences of the segment encompassing the deletion breakpoint were identical in all 10 samples with the −α4.2 deletion. There were four highly conserved single bases (nt 1093 C, nt 1099 C, nt 1121 A, and a C deletion between nt 1140 and nt 1141 positions in the 1596 bp segment) in the −α4.2 allele breakpoint region (registered on GeneBank; accession number AF221717), which were obviously diverse from the corresponding nucleotide positions on the two normal homologous segments (nt 31227 C, nt 31233 T, nt 31255 G, and nt 31275 C in X box 2; nt 35483 T, nt 35489 C, nt 35511 G, and a C deletion between nt 35530 and nt 35531 in X box 1; nucleotide positions from NG_000006). Figure 2 summarises these nucleotide variations. No sequence difference was found in the homologous segments involving two X boxes from the four normal Chinese individuals, and the sequences obtained from these four samples were identical to the sequence data from NG_000006. The DHPLC assay was subsequently designed to distinguish between the normal and the −α4.2 allele by exploiting the four nucleotide differences at the breakpoint region.
Figure 2.
Alignment of the −α4.2 breakpoint region sequence and its homologous normal sequences in Chinese individuals and the data from NG_000006. Four single base variations are marked with open black boxes. X42, the sequence of the −α4.2 allelic breakpoint region from Chinese −α4.2 thalassaemia carriers (nt 1091–1148 in the 1596 bp amplified segment); NGX2 and NGX1 are the homologous sequences representing the X2 box (nt 31225–31283) and the X1 box (nt 35481–35538), respectively, from NG_000006; CNX2 and CNX1 are the homologous sequences representing the X2 box (nt 1091–1149) and the X1 box (nt 1023–1080), respectively, from the sequenced 1302 bp and 1519 bp segments of the normal Chinese individuals
Take home messages.
We have developed a polymerase chain reaction (PCR)/denaturing high performance liquid chromatography (DHPLC) technique that can distinguish the three major genotypes necessary for genotyping the −α4.2 thalassaemia deletion (−α4.2/αα, −α4.2/--SEA, and αα/αα)
This technique showed 100% (40 of 40) concordance with the genotypes previously characterised by Southern blotting or Gap-PCR and was simple, rapid, and cost effective
The PCR/DHPLC approach could be adapted for use in epidemiological surveys, genetic screening, and diagnosis of silent α+ thalassaemia and Hb H disease
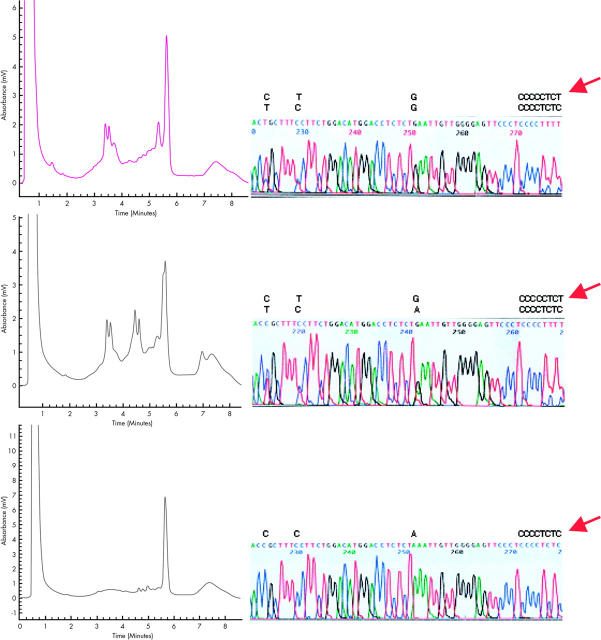
Samples with the −α4.2/αα, −α4.2/--SEA, and αα/αα genotypes could be clearly distinguished by the DHPLC chromatograms (fig 3). Samples with the −α4.2/--SEA genotype showed a single peak. This peak is representative of the −α4.2 allele because the corresponding –SEA allele lacked the segment involving the two X boxes.2,17 In contrast, the −α4.2/αα and αα/αα alleles revealed distinct DHPLC profiles. The chromatographic peaks generated from these genotypes are characterised by multiple double peaks. The double peaks are indicative of the presence of heteroduplexes, which are formed as the result of the presence of the nucleotide differences between the breakpoint region and the corresponding normal homologous regions (fig 3).
Figure 3.
The representative denaturing high performance liquid chromatography (DHPLC) profiles for genotyping of the −α4.2 deletion. WAVE elution profiles (left) and the corresponding sequencing results (right) of the samples with the three major genotypes: (A) αα/αα, (B) −α4.2/αα, and (C) −α4.2/--SEA. The three genotypes can be discriminated by their specific elution peak patterns. There are three and four double peaks formed in the αα/αα and −α4.2/αα samples, respectively, whereas the −α4.2/--SEA samples were characterised by a single peak. The chromatographic profile is determined by a combination of heteroduplexes and homoduplexes formed between the alignment of complementary sequences between the 312 bp amplicons of the X1 and X2 boxes and the −α4.2 breakpoint region. The polymerase chain reaction amplicons were generated with primers D1 and D2 and the sequence differences between the amplicons were confirmed by direct sequencing, as shown. The four single nucleotide polymorphisms within the X boxes characterised in our study are shown above each sequencing trace. The heterozygous sites resulted in the formation of heteroduplexes that are resolved as double peaks on the DHPLC chromatogram.
The specificity and reliability of this newly developed DHPLC approach was compared with that of Gap-PCR or Southern blot analyses in a blind manner. The assay was performed on a total of 40 blood and prenatal DNA archival samples previously characterised by Gap-PCR or Southern blotting. The Gap-PCR or Southern blotting results were unknown until the genotypes obtained by DHPLC were scored. All of the 40 samples tested by DHPLC were found to be 100% concordant with the data obtained by the other two methods.
DISCUSSION
We have sequenced the breakpoint region of the −α4.2 allele and its normal homologous counterparts in the human α globin gene cluster from Chinese individuals. Interestingly, direct sequencing analysis of the −α4.2 deletion junction revealed four single nucleotide polymorphisms. We think that this variation is a conservative genetic event because these polymorphisms were present in all of the 10 −α4.2 gene carriers initially studied. The consistent presence of this polymorphic haplotype was confirmed by subsequent sequence analysis of the samples correctly typed by the novel DHPLC assay (fig 3). As shown in fig 2, the unique sequence haplotype linked to the −α4.2 allele was consistently different from the normal allele sequence haplotypes involving both the X2 and X1 boxes. This four nucleotide polymorphism provided a useful means for the development of new diagnostic assays for the direct detection of the −α4.2 allele in Chinese populations.
DHPLC is a rapid, accurate, and semi-automated technique optimised for the screening of single base mutations, small DNA deletions, and insertions. The heteroduplex and homoduplex DNA fragments formed as a result of the presence or absence of sequence differences between complementary DNA fragments can be discriminated on the transgenomic DHPLC analyser under partial denaturing conditions.13,18 Based on the novel sequence information on the four nucleotide polymorphic haplotypes linked to the −α4.2 allele in our study, we designed a PCR/DHPLC based method for the rapid genotyping of the −α4.2 thalassaemia deletion. As shown in fig 3, the results showed that the three major genotypes (−α4.2/αα, −α4.2/--SEA, and αα/αα) could be easily distinguished based on their respective DHPLC profiles. The peak patterns were related to the formation of heteroduplexes and homoduplexes between the 312 bp amplicons of the X boxes and the breakpoint junction of the −α4.2 allele. Because of the unavailability of the relevant clinical samples, our study did not include the homozygous −α4.2 deletion.
“The high throughput would enable this technique to be adapted for large scale population screening”
The results obtained by the DHPLC approach were 100% accurate for the identification of the −α4.2 deletion when compared with Southern blotting or Gap-PCR. The DHPLC approach enables the use of a single genomic template amplified in a single PCR reaction, with no need for labels for the accurate detection of the −α4.2 mutation. This represents a considerable reduction in labour and costs. The data generated by the WAVE system are easily interpretable and results are generated automatically within seven minutes. The high throughput would enable this technique to be adapted for large scale population screening.14 The PCR/DHPLC technology described in our study was designed for the specific detection of the unique polymorphic haplotype associated with the Chinese −α4.2 allele. We plan to extend this approach to other ethnic populations where the α4.2 deletion is also prevalent by first determining the sequence around the −α4.2 breakpoint region in such populations. Our preliminary results obtained from a few −α4.2 carriers of Middle Eastern origin revealed other unique sequence variations around the breakpoint region. Additional −α4.2 thalassaemia alleles will be analysed in future studies. These sequence data are not only useful for the development of molecular diagnostic approaches, but also provide a useful means for the study of human molecular evolution and genetic recombination mechanisms.19,20
Our study indicates that the −α4.2 deletion is one of the most common α thalassaemia mutations in southern China, with a prevalence of approximately 1.0%, ranking third after the --SEA/and −α3.7 mutations. Consequently, Hb H disease is highly prevalent in southern China and poses a public health concern. Because of a lack of phenotypic features that are characteristic of α+ thalassaemia, molecular genotyping is needed for the epidemiological survey, genetic screening, and diagnosis of α+ thalassaemia carriers and individuals with Hb H disease. These aims could readily be achieved through the adoption of the new technology described here, which is well suited for the rapid and accurate genotyping of the −α4.2 deletion.
Acknowledgments
We thank Dr R Chiu for proof reading the manuscript and invaluable suggestions. This study was partially funded by National Science and Technology Department of China for the key project grants (001CB510308) and by PLA grants for outstanding young scientist in medical research (01J010, to X-M Xu).
Abbreviations
DHPLC, denaturing high performance liquid chromatography
NCBI, National Center for Biotechnology Information
PCR, polymerase chain reaction
TEAA, triethylammonium acetate
REFERENCES
- 1.Lauer J, Shen CK, Maniatis T. The chromosomal arrangement of human alpha-like globin genes: sequence homology and alpha-globin gene deletions. Cell 1980;20:119–30. [DOI] [PubMed] [Google Scholar]
- 2.Higgs DR, Vickers MA, Wilkie AO, et al. A review of the molecular genetics of the human alpha-globin gene cluster. Blood 1989;73:1081–104. [PubMed] [Google Scholar]
- 3.Yenchitsomanus PT, Summers KM, Bhatia KK, et al. Extremely high frequencies of alpha-globin gene deletion in Madang and on Kar Kar Island, Papua New Guinea. Am J Hum Genet 1985;37:778–84. [PMC free article] [PubMed] [Google Scholar]
- 4.Flint J, Harding RM, Boyce AJ, et al. The population genetics of the haemoglobinopathies. Baillieres Clin Haematol 1998;11:1–51. [DOI] [PubMed] [Google Scholar]
- 5.Weatherall DJ. Phenotype–genotype relationships in monogenic disease: lessons from the thalassaemias. Nat Rev Genet 2001;2:245–55. [DOI] [PubMed] [Google Scholar]
- 6.Chui DH, Fucharoen S, Chan V. Hemoglobin H disease: not necessarily a benign disorder. Blood 2003;101:791–800. [DOI] [PubMed] [Google Scholar]
- 7.Embury SH, Miller JA, Dozy AM, et al. Two different molecular organizations account for the single alpha-globin gene of the alpha-thalassemia-2 genotype. J Clin Invest 1980;66:1319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oron-Karni V, Filon D, Oppenheim A, et al. Rapid detection of the common Mediterranean alpha-globin deletions/rearrangements using PCR. Am J Hematol 1998;58:306–10. [DOI] [PubMed] [Google Scholar]
- 9.Chong SS, Boehm CD, Higgs DR, et al. Single-tube multiplex-PCR screen for common deletional determinants of alpha-thalassemia. Blood 2000;95:360–2. [PubMed] [Google Scholar]
- 10.Chu DC, Lee CH, Lo MD, et al. Non-radioactive Southern hybridization for early diagnosis of alpha-thalassemia with southeast Asian-type deletion in Taiwan. Am J Med Genet 2000;95:332–5. [PubMed] [Google Scholar]
- 11.Liu YT, Old JM, Miles K, et al. Rapid detection of alpha-thalassaemia deletions and alpha-globin gene triplication by multiplex polymerase chain reactions. Br J Haematol 2000;108:295–9. [DOI] [PubMed] [Google Scholar]
- 12.Fucharoen S, Winichagoon P, Wisedpanichkij R, et al. Prenatal and postnatal diagnoses of thalassemias and hemoglobinopathies by HPLC. Clin Chem 1998;44:740–8. [PubMed] [Google Scholar]
- 13.Kuklin A, Munson K, Gjerde D, et al. Detection of single-nucleotide polymorphisms with the WAVE DNA fragment analysis system. Genet Test 1997–98;1:201–6. [DOI] [PubMed] [Google Scholar]
- 14.Xiao W, Oefner PJ. Denaturing high-performance liquid chromatography: a review. Hum Mutat 2001;17:439–74. [DOI] [PubMed] [Google Scholar]
- 15.Liebhaber SA, Goossens M, Kan YW. Homology and concerted evolution at the alpha 1 and alpha 2 loci of human alpha-globin. Nature 1981;290:26–9. [DOI] [PubMed] [Google Scholar]
- 16.Hess JF, Fox M, Schmid C, et al. Molecular evolution of the human adult alpha-globin-like gene region: insertion and deletion of Alu family repeats and non-Alu DNA sequences. Proc Natl Acad Sci U S A 1983;80:5970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winichagoon P, Higgs DR, Goodbourn SE, et al. The molecular basis of alpha-thalassaemia in Thailand. EMBO J 1984;3:1813–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinka T, Naroda T, Tamura T, et al. A rapid and simple method for sex identification by heteroduplex analysis, using denaturing high-performance liquid chromatography (DHPLC). J Hum Genet 2001;46:263–6. [DOI] [PubMed] [Google Scholar]
- 19.Fodde R, Losekoot M, Casula L, et al. Nucleotide sequence of the Belgian G gamma+ (A gamma delta beta)0-thalassemia deletion breakpoint suggests a common mechanism for a number of such recombination events. Genomics 1990;8:732–5. [DOI] [PubMed] [Google Scholar]
- 20.Currat M, Trabuchet G, Rees D, et al. Molecular analysis of the beta-globin gene cluster in the Niokholo Mandenka population reveals a recent origin of the beta(S) Senegal mutation. Am J Hum Genet 2002;70:207–23. [DOI] [PMC free article] [PubMed] [Google Scholar]