Abstract
Aims: Use of the polymerase chain reaction for the detection of Mycobacterium tuberculosis (TB PCR) as a basis for making clinical decisions on the initiation of antituberculosis treatment was studied.
Methods: A retrospective study involving a cohort of 155 patients being investigated for tuberculosis in an infectious disease consultation service was undertaken. TB PCR was performed on pulmonary and extrapulmonary specimens from these patients. The sensitivity of TB PCR was analysed.
Results: Of the 155 patients, 144 fitted the clinical diagnosis of tuberculosis, and 112 of them were culture positive for M tuberculosis. Sixty (58.3%) patients with clinical features suggestive of tuberculosis received antituberculosis treatment based on positive TB PCR alone. Of 224 clinical specimens (138 pulmonary and 86 extrapulmonary) sent for TB PCR, 148 (99 pulmonary and 49 extrapulmonary) were positive in 117 patients. Of the 690 clinical specimens sent for culture, 279 were positive for M tuberculosis in 112 patients. The diagnostic sensitivity of TB PCR was 75.9% (85 of 112) and 81.3% (117 of 144) in patients with culture confirmed and clinically diagnosed tuberculosis, respectively. Using culture as the gold standard, the overall sensitivity of TB PCR was 78.3%, and for pulmonary and extrapulmonary specimens it was 82.3% and 72.0%, respectively.
Conclusions: TB PCR is a rapid and reliable test in the diagnosis and management of tuberculosis.
Keywords: polymerase chain reaction, Mycobacterium tuberculosis, clinical evaluation
In clinical practice, antituberculosis treatment is often started purely on the basis of compatible clinical symptoms and signs, suggestive radiological changes, and a rapid microbiological test result, either a positive smear for acid fast bacilli (AFB), and/or positive polymerase chain reaction for tuberculosis (TB PCR). Confirmation of tuberculosis by positive culture often takes three to six weeks. Even before the availability of culture results, patients may have clinical deterioration, which could be related to an alternative non-tuberculosis infection, side effects of antituberculosis treatment, antituberculosis drug resistance, or a paradoxical response. The results of TB PCR testing can help to speed up the decision making process involved in the diagnosis of tuberculosis, so that early antituberculosis treatment can be initiated. The laboratory performance of both commercial and in house TB PCR in the diagnosis of tuberculosis has been extensively evaluated.1–9 However, there are few data on the “in use” evaluation of TB PCR as a tool for clinical decision making in the management of tuberculosis.
“Confirmation of tuberculosis by positive culture often takes three to six weeks”
In our study, we report the management of a cohort of 155 patients with clinically suspected tuberculosis. We examined the role of TB PCR using a manual, in house, single tube, nested PCR of the IS6110 gene for the detection of Mycobacterium tuberculosis as a rapid laboratory diagnostic test in the initiation of antituberculosis treatment.
METHODS
Patients
Our study was performed at Queen Mary Hospital, a tertiary hospital with a 1350 bed teaching hospital in Hong Kong, from 1 July 2000 to 30 June 2002. During the study period, all patients referred for infectious disease consultation with clinical features suggestive of tuberculosis were recruited into our study.
Microbiological investigation for M tuberculosis
We performed investigations for M tuberculosis, including AFB smear examination and culture, on relevant clinical specimens of all patients with suspected tuberculosis. TB PCR was performed once a week on pulmonary and extrapulmonary specimens in the following clinical settings: (1) patients with typical radiological changes compatible with pulmonary tuberculosis; (2) patients with radiological changes of old tuberculosis but presenting with new pulmonary infiltrates; (3) patients with community acquired pneumonia not responding to appropriate antibiotic treatment; and (4) patients undergoing invasive procedures, such as image guided or surgical drainage and biopsy for sterile sites in the investigation of extrapulmonary tuberculosis. Histological examination for M tuberculosis was also performed if clinically indicated.
Microbiological processing of clinical specimens
All specimens were collected in γ irradiated disposable plastic containers. A direct smear was prepared for the auramine fluorescent stain and, if positive, the Ziehl-Neelsen stain was performed for confirmation. The respiratory specimens were decontaminated and digested by treatment with an equal volume of sputolysin/sodium hydroxide (4%) for 30 minutes at room temperature with rocking. After neutralisation with 20 ml of 0.067M sodium phosphate buffer (pH 5.3), the mixture was centrifuged at 2750 ×g for 30 minutes. After discarding the supernatant, 400–500 μl of the sediment was obtained. For early morning urine, 20 ml samples were centrifuged at 2750 ×g for 30 minutes. The sediments were digested with 2% sodium hydroxide, followed by neutralisation with 0.067M phosphate buffer (pH 5.3). After centrifugation, sediments were obtained. For the other non-respiratory samples, such as sterile body fluid or tissue aspirates, specimens were microcentrifuged at 12 000 ×g for 10 minutes to collect the sediments. For sediments of both respiratory and non-respiratory specimens, about 60 μl of the sediment was inoculated into two Lowenstein-Jensen medium culture bottles and incubated at 37°C for up to eight weeks. Solid medium slants were considered positive when visible colonies grew. The colonies were further confirmed as mycobacteria by the Ziehl-Neelsen stain. Cultures positive for AFB were identified by the AccuProbe hybridisation assay (Gen-Probe, San Diego, California, USA), according to the manufacturer’s instructions. For mycobacteria other than M tuberculosis, conventional biochemical tests were performed for identification.
The rest of the sediment was transferred to an Eppendorf tube and stored at −20°C if not immediately processed.
Manual one tube nested PCR for the IS6110 gene sequence for the detection of M tuberculosis
For the digested sediments, the Roche Cobas Amplicor (Roche Diagnostics, Berlin, Germany) extraction protocol was used, as described previously.8,9 A volume of 500 μl wash solution was mixed with the sediment. After centrifugation at 15 000 ×g for 10 minutes the supernatant was decanted, 100 μl of lysis reagent was added, and the mixture was vortexed, followed by incubation at 60°C for 45 minutes. Extraction was terminated by adding 100 μl of neutralisation reagent. Each PCR reaction contained 10 μl of the DNA extract. The two sets of primers used for amplification were derived from the gene sequence encoding the insertion sequence IS6110 as follows: external primers were derived from position 367 to 392 (5′-CCGGCCAGCACGCTAATTAACGGTTC-3′) and position 769 to 746 (5′-TGTGGCCGGATCAGCGATCGTGGT-3′); and internal primers were derived from position 455 to 472 (5′-CTGCACACAGCTGACCGA-3′) and position 670 to 652 (5′-CGTTCGACGGTGCATCTG-3′). The reaction mixture consisted of 10mM Tris/HCl (pH 8.3), 50mM KCl, 2mM MgCl2, 0.15mM dATP, dGTP, dCTP, 0.45mM dUTP, 2 pmol external primers, 75 pmol internal primers, 2 U AmpliTaq Gold polymerase (Perkin Elmer, Foster City, California, USA), and 0.5 U heat labile uracil-N-glycosylase (Roche Diagnostics) in a total reaction volume of 100 μl. The mixture was first incubated at 37°C for 10 minutes with uracil-N-glycosylase to destroy any contaminating amplicons, and then at 94°C for 12 minutes to activate the AmpliTaq polymerase, followed by subsequent temperature cycling at 94°C for 45 seconds and 72°C for 1.5 minutes for the first 15 cycles, followed by 94°C for 45 seconds, 55°C for 45 seconds, and 72°C for one minute for 45 cycles. Positive and negative controls were included in each run and all precautions to prevent cross contamination were observed. Amplified products were electrophoresed through a 2% agarose gel in Tris borate buffer. Target bands of 21 bp were visualised by staining with ethidium bromide.
Diagnosis and treatment of M tuberculosis
Preliminary diagnosis of M tuberculosis was based on a positive AFB smear, TB PCR, or tissue histology results during the initial investigation. A definitive diagnosis of tuberculosis was made by positive culture results. For those patients with negative culture for M tuberculosis, a clinical diagnosis was made by an investigators’ consensus with respect to the clinical and radiological presentation and progression, tuberculin skin tests, haematological findings, histological findings (when available), and clinical response to antituberculosis treatment.
Antituberculosis treatment was given to patients with a preliminary or definitive diagnosis of tuberculosis, or empirically if clinical suspicion of M tuberculosis was high.
RESULTS
Patients
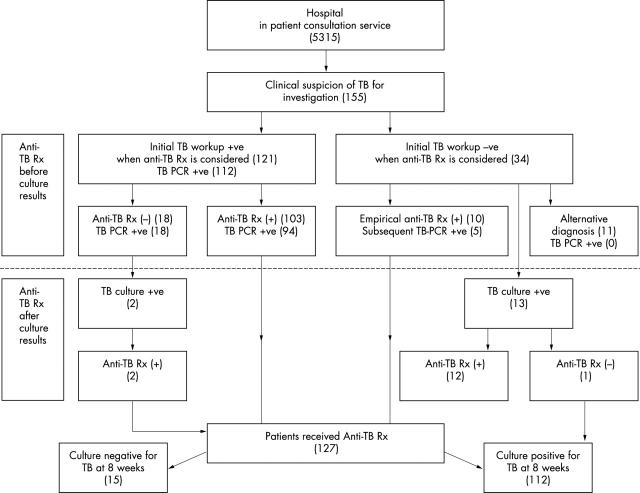
During the two year study period, there were 5315 inpatient infectious disease consultations from various clinical specialties. One hundred and fifty five patients with clinical symptoms and signs suggestive of tuberculosis were recruited into our study and managed according to fig 1. Definitive diagnosis of tuberculosis was documented in 112 patients by positive culture for M tuberculosis. Another 32 patients were diagnosed as having tuberculosis based on clinical consensus among the investigators. Of these 144 patients, 94 had pulmonary tuberculosis, 31 had extrapulmonary tuberculosis, and 19 had combined pulmonary and extrapulmonary tuberculosis. The remaining 11 patients were determined not to have tuberculosis and alternative diagnoses were made.
Figure 1.
Decision for initiating antituberculosis treatment (anti-TB Rx) in 155 patients based on clinical and laboratory findings. TB PCR, polymerase chain reaction for Mycobacterium tuberculosis.
Microbiological investigation for M tuberculosis
In total, 437 pulmonary specimens (sputum, bronchial aspirates, bronchoalveolar lavage, and gastric aspirates) and 253 extrapulmonary (cerebrospinal fluid, pleural effusion, peritoneal fluid, joint fluid, bone marrow aspiration, bone biopsy, tissue biopsy, lymph nodes, urine, and stools) specimens were obtained for AFB smear and culture examinations in 155 patients. Eighty eight smears were positive in 35 patients and 279 cultures were positive for M tuberculosis in 112 patients, respectively. The diagnostic sensitivity of the AFB smear was 31.3% (35 of 112) and 24.3% (35 of 144) in culture confirmed and clinically diagnosed tuberculosis, respectively. The yield from AFB culture was 77.8% when clinical diagnosis was used as reference.
In total, 224 clinical specimens were requested from 155 patients for TB PCR (138 pulmonary specimens and 86 extrapulmonary specimens), in which 148 specimens (99 pulmonary specimens and 49 extrapulmonary specimens) were positive in 117 patients. Figure 2 shows the correlations between TB PCR and AFB smear and culture results. One sample in one patient with a positive AFB smear who was culture positive for M tuberculosis showed negative results in the TB PCR test. A PCR inhibitor was detected in the sample extract from this patient. Eight patients with histological features of tuberculosis but who were negative in TB PCR had positive M tuberculosis cultures. Thirty four patients with a positive smear and TB PCR had a positive culture for M tuberculosis. Another 83 patients had a negative smear and a positive TB PCR in the initial investigation, but culture was subsequently positive in 51 patients. Those patients with a positive TB PCR but a negative culture had clinical and radiological results that were suggestive of active tuberculosis. TB PCR was positive in 85 of 112 patients who were culture positive for M tuberculosis. The diagnostic sensitivity of TB PCR was 75.9% (85 of 112) and 81.3% (117 of 144) in culture confirmed and clinically diagnosed tuberculosis, respectively. TB PCR was positive in 101 of the 129 specimens (79 pulmonary specimens and 50 extrapulmonary specimens) where cultures were positive. The overall sensitivity of the assay was 78.3%, and that of pulmonary and extrapulmonary specimens was 82.3% and 72.0%, respectively (table 1). In the 11 patients in whom an alternative diagnosis other than tuberculosis was made, 54 and 11 clinical specimens were negative for mycobacterial culture and TB PCR, respectively, resulting 100% specificity of TP PCR. Tissue specimens were available in 20 patients for histological examination and 12 of them had evidence of mycobacterial infection.
Figure 2.
The correlation between polymerase chain reaction for Mycobacterium tuberculosis (TB PCR) and conventional work up for M tuberculosis in 155 patients. *Those patients with positive TB PCR but negative culture results had clinical and radiological results that were suggestive of active tuberculosis.
Table 1.
Evaluation of TB PCR in culture confirmed Mycobacterium tuberculosis
| Total number of culture positive specimens | Number of TB PCR positive samples | Number of TB PCR negative samples | Sensitivity of TB PCR | |
| Pulmonary specimens | 79 | 65 | 14 | 82.3% |
| Sputum | 39 | 31 | 8 | 79.5% |
| Bronchoalveolar lavage | 40 | 34 | 6 | 85% |
| Extrapulmonary specimens | 50 | 36 | 14 | 72% |
| Tissue | 16 | 13 | 3 | 81.2% |
| Aspirates | 13 | 9 | 4 | 69.2% |
| Cerebrospinal fluid | 10 | 8 | 2 | 80% |
| Pleural fluid | 6 | 2 | 4 | 33.3% |
| Early morning urine | 4 | 3 | 1 | 75% |
| Endocervical swab | 1 | 1 | 0 | 100% |
| Total | 129 | 101 | 28 | 78.3% |
TB PCR, polymerase chain reaction for Mycobacterium tuberculosis.
Diagnosis and treatment of M tuberculosis
In the initial investigation, 121 patients had laboratory evidence of M tuberculosis infection, which included AFB smear and TB PCR positive in 34, AFB smear positive but TB PCR negative in one, AFB smear negative but TB PCR positive in 78, and histological features of mycobacterial infection in eight patients. Standard antituberculosis treatment including isoniazid, rifampicin, pyrazinamide, and ethambutol was given to 103 patients when the initial work up results became available. Thirty four (33%) of 103 patients were treated because of positive smear and TB PCR results, and 60 (58.3%) patients received antituberculosis therapy based on positive TB PCR results. Treatment was started because of a positive AFB smear result in one and a positive histological result alone in eight patients. Antituberculosis treatment was not given in 18 patients because of severe comorbidity in 15, an advanced age of 102 in one, and the patient’s refusal for fear of potential side effects of treatment in two, despite positive TB PCR results at the initial investigation for TB. In 34 patients with negative AFB smears, empirical antituberculosis treatment was given as above while waiting for the TB PCR results in 10 (6.9%) patients. There was pulmonary or miliary tuberculosis in six patients, tuberculous meningitis in three, and tuberculous spondylitis and peritonitis in one. Five of these 10 patients turned out to have a positive TB PCR within five days of starting treatment. The remaining 24 patients with negative results in both the AFB smear and TB PCR were investigated further and an alternative diagnosis was made in 11 patients. Fourteen patients received antituberculosis treatment when the culture results were available, including two patients who refused initial treatment. Therefore, 127 (82%) of 155 patients received antituberculosis treatment in this cohort, of whom 118 completed the course of treatment. Nine patients defaulted follow up after the initiation of treatment.
DISCUSSION
The decision to initiate antituberculosis treatment based on the pretreatment nucleic acid amplification assay on both respiratory and non-respiratory specimens was studied. In our series, of 144 patients with a clinical diagnosis of M tuberculosis infection, only 25% had positive results on AFB smear examination, but about 80% had a positive TB PCR assay result. Almost 60% patients received antituberculosis treatment based on positive TB PCR as the sole result in the initial investigation, and more than 70% of them had subsequent culture confirmation of M tuberculosis. Without this rapid assay, delay in giving the appropriate treatment would probably have occurred.
Although the Food and Drug Administration of the USA recommended that PCR should only be performed for rapid diagnosis in respiratory specimens of either AFB smear positive or negative samples,10 nucleic acid amplification has been studied extensively in patients with extrapulmonary tuberculosis using either in house or commercial kits. The sensitivity ranged between 42% and 93% in culture positive specimens.6,11–15 Our centre uses a manual in house, single tube, nested PCR using IS6110 as a target site for the rapid diagnosis of pulmonary and extrapulmonary tuberculosis,5–7 with a specificity of 99.7%.6,7 The manual assay also showed comparable results to the commercial kits when respiratory specimens were evaluated.8,9 In our study, the overall sensitivity of the TB PCR assay was 78.3%, and 72% for extrapulmonary specimens, which is similar to that seen in another centre using IS6110 as a target site for laboratory diagnosis.4 However, the result for pleural fluid in our study was particularly poor, with a sensitivity of only 33.3%, in contrast to another study, which found the sensitivity of PCR to be 81% for pleural fluid.16 The presence of inhibitors could not explain the discrepancy in these results. Interestingly, we found a positive TB PCR in an endocervical swab, which has not been reported in the literature.
“Of 144 patients with a clinical diagnosis of Mycobacterium tuberculosis infection, only 25% had positive results on acid fast bacilli smear examination, but about 80% had a positive Mycobacterium tuberculosis polymerase chain reaction result”
Although TB PCR appears to be a rapid, sensitive, and specific diagnostic assay, the results should be interpreted with care in the clinical setting. In our study, 32 patients had a positive TB PCR but a negative culture subsequently. In such cases, there is a clinical dilemma of whether antituberculosis treatment should be maintained or discontinued. As long as precautions have been taken to avoid cross contamination, the positive TB PCR may indicate the presence of non-viable AFB, particularly in patients with a history of tuberculosis in the past. However, a negative AFB culture cannot preclude a clinical diagnosis of tuberculosis, particularly in patients with clinical and radiological features suggestive of active tuberculosis, as demonstrated in our patients. There can be a variety of reasons for a positive TB PCR but negative AFB culture. For example, the M tuberculosis count may be low at the tissue level or the organisms may not be viable as a result of the decontamination procedure used when handling respiratory specimens.17 Therefore, TB PCR can be used to complement culture in selected patients.
Take home messages.
The polymerase chain reaction for Mycobacterium tuberculosis (TB PCR) is a rapid and reliable method for the diagnosis of both pulmonary and extrapulmonary tuberculosis, with an overall sensitivity of 78.3% and a specificity of 100%
It enabled antituberculosis treatment to be started early in more than half of the patients studied
Because of the relatively low sensitivity of TB PCR, clinical judgment remains the ultimate decision in the management of tuberculosis
Recent advances in the molecular diagnosis of tuberculosis involving the detection of bacterial mRNA encoding the ubiquitous 85B antigen in viable M tuberculosis by reverse transcriptase PCR and reverse transcriptase strand displacement amplification assay may overcome the above difficulty.18–20 Because the mRNA is more rapidly destroyed in the cells than rRNA or genomic DNA, having a half life of only a few minutes, it can differentiate between viable and non-viable M tuberculosis. Thus, it may become useful for the diagnosis of active tuberculosis and provide a tool to monitor the therapeutic efficacy.
In conclusion, TB PCR is a rapid and reliable method in the diagnosis of tuberculosis, which allowed early initiation of antituberculosis treatment in more than 50% of the studied patients in our cohort. However, because of the relatively low sensitivity of TB PCR, clinical judgment remains the ultimate decision in the management of tuberculosis.
Abbreviations
AFB, acid fast bacilli
PCR, polymerase chain reaction
TB, Mycobacterium tuberculosis
REFERENCES
- 1.Brown TJ, Power EG, French GL. Evaluation of three commercial detection systems for Mycobacterium tuberculosis where clinical diagnosis is difficult. J Clin Pathol 1999;52:193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajalahti I, Vuorinen P, Liippo K, et al. Evaluation of commercial DNA and rRNA amplification assays for assessment of treatment outcome in pulmonary tuberculosis patients. Eur J Clin Microbiol Infect Dis 2001;20:746–50. [DOI] [PubMed] [Google Scholar]
- 3.Bogard M, Vincelette J, Antinozzi R, et al. Multicenter study of a commercial, automated polymerase chain reaction system for the rapid detection of Mycobacterium tuberculosis in respiratory specimens in routine clinical practice. Eur J Clin Microbiol Infect Dis 2001;20:724–31. [DOI] [PubMed] [Google Scholar]
- 4.Almeda J, Garcia A, Gonzalez J, et al. Clinical evaluation of an in-house IS6110 polymerase chain reaction for diagnosis of tuberculosis. Eur J Clin Microbiol Infect Dis 2000;19:859–67. [DOI] [PubMed] [Google Scholar]
- 5.Yuen KY, Chan KS, Chan CM, et al. Use of PCR in routine diagnosis of treated and untreated pulmonary tuberculosis. J Clin Pathol 1993;46:318–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan CM, Yuen KY, Chan KS, et al. Single-tube nested PCR in the diagnosis of tuberculosis. J Clin Pathol 1996;49:290–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuen KY, Chan KS, Chan CM, et al. Monitoring the therapy of pulmonary tuberculosis by nested polymerase chain reaction assay. J Infect 1997;34:29–33. [DOI] [PubMed] [Google Scholar]
- 8.Yuen KY, Yam WC, Wong LP, et al. Comparison of two automated DNA amplification systems with a manual one-tube nested PCR assay for diagnosis of pulmonary tuberculosis. J Clin Microbiol 1997;35:1385–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yam WC, Yuen KY, Seto WH. Direct detection of Mycobacterium tuberculosis in respiratory specimens using an automated DNA amplification assay and a single tube nested polymerase chain reaction (PCR). Clin Chem Lab Med 1998;36:597–9. [DOI] [PubMed] [Google Scholar]
- 10.Diagnostic standards and classification of tuberculosis in adults and children. Am J Respir Crit Care Med 2000;161(4 Pt 1):1376–95. [DOI] [PubMed] [Google Scholar]
- 11.Kearns AM, Freeman R, Steward M, et al. A rapid polymerase chain reaction technique for detecting Mycobacterium tuberculosis in a variety of clinical specimens. J Clin Pathol 1998;51:922–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolk AH, Kox LF, van Leeuwen J, et al. Clinical utility of the polymerase chain reaction in the diagnosis of extrapulmonary tuberculosis. Eur Respir J 1998;11:1222–6. [DOI] [PubMed] [Google Scholar]
- 13.Bemer-Melchior P, Germaud P, Drugeon HB. Diagnosis of extrapulmonary tuberculosis by a commercial polymerase chain reaction kit. Pathol Biol (Paris) 1998;46:597–603. [PubMed] [Google Scholar]
- 14.Tortoli E, Tronci M, Tosi CP, et al. Multicenter evaluation of two commercial amplification kits (Amplicor, Roche and LCx, Abbott) for direct detection of Mycobacterium tuberculosis in pulmonary and extrapulmonary specimens. Diagn Microbiol Infect Dis 1999;33:173–9. [DOI] [PubMed] [Google Scholar]
- 15.Portillo-Gomez L, Morris SL, Panduro A. Rapid and efficient detection of extra-pulmonary Mycobacterium tuberculosis by PCR analysis. Int J Tuberc Lung Dis 2000;4:361–70. [PubMed] [Google Scholar]
- 16.Querol JM, Minguez J, Garcia-Sanchez E, et al. Rapid diagnosis of pleural tuberculosis by polymerase chain reaction. Am J Respir Crit Care Med 1995;152(6 Pt 1):1977–81. [DOI] [PubMed] [Google Scholar]
- 17.Metchock BG, Nolte FS, Wallace RJ Jr. Mycobacterium. In: Murray PR, Baron EJ, Pfaller MA, et al, eds. Manual of clinical microbiology, 7th ed. Washington DC: American Society for Microbiology, 1999.
- 18.Jou NT, Yoshimori RB, Mason GR, et al. Single-tube, nested, reverse transcriptase PCR for detection of viable Mycobacterium tuberculosis. J Clin Microbiol 1997;35:1161–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellyer TJ, DesJardin LE, Hehman GL, et al. Quantitative analysis of mRNA as a marker for viability of Mycobacterium tuberculosis. J Clin Microbiol 1999;37:290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hellyer TJ, DesJardin LE, Teixeira L, et al. Detection of viable Mycobacterium tuberculosis by reverse transcriptase-strand displacement amplification of mRNA. J Clin Microbiol 1999;37:518–23. [DOI] [PMC free article] [PubMed] [Google Scholar]