Abstract
Aim: Runx transcription factors are important regulators of lineage specific gene expression, cell proliferation, and differentiation. Runx3 expression is lost in a high proportion of gastric cancers, suggesting a tumour suppressive role in this malignancy. This study investigates the expression and localisation of Runx3 in pancreatic tissues.
Methods: Quantitative polymerase chain reaction was used to measure Runx3 mRNA. Immunohistochemistry was carried out to localise Runx3 in normal pancreatic tissues, and in primary and metastatic pancreatic ductal adenocarcinoma (PDAC). Basal and transforming growth factor β1 (TGFβ1) induced Runx3 expression was analysed in cultured pancreatic cancer cell lines.
Results: Runx3 expression was low to absent in normal pancreatic tissues, but increased in a third of cancer tissues. Runx3 was present only in islets in normal pancreas, whereas in pancreatic cancers, Runx3 was detected in the cancer cells of seven of 24 samples analysed. In addition, it was expressed by lymphocytes in six of the 16 cases with lymphocyte infiltration. In pancreatic cancer cell lines, Runx3 mRNA was present in Colo-357 and T3M4 cells, but was low to absent in the other cell lines tested. TGFβ1 repressed Runx3 mRNA expressed in Colo-357 cells, and had no effect on Runx3 expression in the other pancreatic cancer cell lines.
Conclusion: Runx3 expression is restricted to islets in the normal pancreas. In contrast, a considerable proportion of pancreatic tumours express Runx3, and its expression is localised in the tumour cells and in the infiltrating lymphocytes. Thus, Runx3 might play a role in the pathogenesis of PDAC.
Keywords: Runx3, transforming growth factor β, pancreatic cancer
Despite its relatively low incidence of approximately 10 cases/100 000 people, pancreatic cancer is still one of the leading causes of cancer related death in Western industrialised countries, as a result of its poor prognosis, with an overall five year survival rate of less than 5%.1 The pathogenesis of pancreatic ductal adenocarcinoma (PDAC) can be described as a step by step accumulation of genetic changes, such as K-ras oncogene mutations, p53, p16, and Smad4 tumour suppressor gene mutations,2 in addition to several epigenetic alterations, which together result in self sufficiency of growth signals, insensitivity to antigrowth signals, evasion of apoptosis, angiogenesis, invasion, and metastasis.3
“Runx proteins can act both as protooncogenes and tumour suppressor genes”
Runx proteins have essential functions in both cell proliferation and differentiation in humans. For example, Runx1 and Runx2 play fundamental roles in haemopoietic and osteogenic lineage specific gene expression.4,5 In addition, they can act both as protooncogenes and tumour suppressor genes. For example, Runx1 mutations are associated with human leukaemia,6 and Runx3 was suggested to be a tumour suppressor gene in gastric cancer, where loss of Runx3 expression is seen in 45–60% of cases.7 Runx3 was initially found to be a neurogenic TrkC neurospecific transcription factor,8 and also has crucial functions in lineage specification and homeostasis of CD8 positive T cells.9 Runx proteins also function as transcriptional regulators of the Smad gene family, which mediates downstream signalling by transforming growth factor β (TGFβ) and its homologues.10 TGFβs are multifunctional cytokines that play important roles in regulating cellular growth and differentiation in many biological systems.8 Smad4 is a key transcription factor in the TGFβ1 signalling pathway, and is inactivated in about 50% of pancreatic adenocarcinomas. Because Runx3 functions as a tumour suppressor gene in gastric cancer, and in view of the important role of TGFβ and Smad proteins in PDAC, the aim of our present study was to investigate the expression and localisation of Runx3 in PDAC, in addition to its role in the TGFβ pathway.
MATERIALS AND METHODS
Cell lines and culture conditions
MiaPaCa-2, T3M4, Aspc-1, Bxpc-3, Capan-1, Colo-357, SU8686, and Panc-1 pancreatic cancer cells were grown in RPMI 1640 medium containing 10% fetal bovine serum, 100 U?ml penicillin, and 100 mg/ml streptomycin (Invitrogen, Karlsruhe, Germany). Cells were maintained at 37°C in humidified air with 5% CO2. For induction experiments, cells were treated with 200pM TGFβ1 (R&D Systems Inc, Minneapolis, Minnesota, USA) for the indicated time.
Patients and tissue collection
Sixty two PDAC samples, including five liver metastases and five lymph node metastases, were obtained from 37 male and 35 female patients (median age, 65 years; range, 28–82) who underwent pancreatic resections because of PDAC at the university hospitals of Berne (Switzerland) and Heidelberg (Germany). Thirty nine human chronic pancreatitis (CP) tissue samples were obtained from 30 male and nine female patients (median age, 54 years; range, 25–73). Twenty four normal human pancreatic tissue samples were obtained from previously healthy individuals (median age, 45 years; range, 20–74) through an organ donor programme. Of the 52 primary PDACs, nine tumours were stage I, six were stage II, 34 were stage III, and three were stage IV according to the 1997 TNM classification of the Union International Contre le Cancer. Fourteen tumours were well differentiated, 27 moderately, and 11 poorly differentiated. Most of the tumours were located in the head of the pancreas (51 cases), and only one tumour was located in the pancreatic tail. Immediately after surgical removal, tissue samples were either snap frozen in liquid nitrogen and then maintained at 80°C until use (for RNA extraction) or fixed in 5% formalin and embedded in paraffin wax after 24 hours. All studies were approved by the ethics committees of the University of Heidelberg and the University of Bern.
Real time quantitative polymerase chain reaction
All reagents and equipment for mRNA and cDNA preparation were purchased from Roche (Roche Applied Science, Mannheim, Germany). mRNA was prepared by automated isolation using the MagNA Pure LC instrument and isolation Kit I (for cells) and Kit II (for tissues). RNA was reverse transcribed into cDNA using the First Strand cDNA synthesis kit for reverse transcription polymerase chain reaction (PCR) (AMV) according to the manufacturer’s instructions. Real time quantitative PCR (QRT-PCR) was performed with the Light Cycler Fast Start DNA SYBR Green kit, as described previously.11 The number of specific transcripts was normalised to housekeeping genes (cyclophilin B and hypoxanthine guanine phosphoribosyltransferase (HPRT)). All primers were obtained from Search-LC (Heidelberg, Germany).
Western blot analysis of pancreatic cell lines
Cells were lysed in a suspension buffer (50mM Tris/HCl, 150mM NaCl, 2mM EDTA, 1% sodium dodecyl sulfate (SDS)) containing complete protease inhibitor cocktail tablets from Roche (Roche Applied Science). Protein concentrations were measured with the BCA protein assay (Pierce Chemical Co, Rockford, Illinois, USA). The cell lysates (30 µg aliquots) were separated on SDS polyacrylamide gels and electroblotted on to nitrocellulose membranes. Membranes were then incubated in blocking solution (5% non-fat milk in 20mM Tris/HCl, 150mM NaCl, 0.1% Tween-20; TBS-T), followed by incubation with rabbit anti-Runx3 antibodies (1/5000 dilution)8 at 4°C overnight. The membranes were then washed in TBS-T and incubated with horseradish peroxidase conjugated secondary antibodies (Amersham Life Science, Amersham, Buckinghamshire, UK) for one hour at room temperature. Antibody detection was performed with an enhanced chemiluminescence reaction (Amersham Life Science).
Immunohistochemistry
Immunohistochemistry was performed using the Dako EnVision Systems (Dako Cytomation GmbH, Hamburg, Germany). Consecutive paraffin wax embedded tissue sections (3–5 µm thick) were dewaxed and rehydrated. Antigen retrieval was performed by pretreatment of the slides in citrate buffer (pH 6.0) in a microwave oven for 12 minutes. Thereafter, slides were cooled to room temperature in deionised water for five minutes. Endogenous peroxidase activity was quenched by incubating the slides in methanol containing 0.6% hydrogen peroxide, followed by washing in deionised water for three minutes, after which the sections were incubated for one hour at room temperature with normal goat serum, and subsequently incubated at 4°C overnight with the primary antibody (anti-Runx3; 1/1000 dilution). Next, the sections were rinsed with washing buffer (TBS with 0.1% bovine serum albumin) and incubated with horseradish peroxidase linked goat antirabbit antibodies, followed by reaction with diaminobenzidine and counterstaining with Mayer’s haematoxylin. The specificity of the anti-Run3 antibody has been established previously.12 In addition, to confirm the specificity of the primary antibody and the technique used here, tissue sections were incubated in the absence of the primary antibody and with negative control rabbit IgG. Under these conditions, no specific immunostaining was detected.
RESULTS
Runx3 expression and localisation in pancreatic tissues
QRT-PCR was performed to evaluate the amounts of Runx3 mRNA in normal pancreatic tissue samples (n = 24), chronic pancreatitis (n = 39), and pancreatic ductal adenocarcinoma samples (n = 52). Tissue samples from normal tissues had a mean (SEM) replicate number of Runx3 mRNA of 6.4 (2.3), whereas Runx3 mRNA was significantly increased in both chronic pancreatitis and PDAC, with the mean (SEM) replicate number being 114.1 (57.3) in chronic pancreatitis (p < 0.05) and 204.2 (128.4) (p < 0.05) in PDAC (fig 1). The difference in Runx3 mRNA values between CP and PDAC was not significant. No correlation was found between Runx3 expression and tumour stage or grade.
Figure 1.

Runx3 mRNA expression in pancreatic tissues: Runx3 mRNA values in normal pancreatic tissues, chronic pancreatitis (CP), and pancreatic ductal adenocarcinoma (cancer) tissues by real time quantitative polymerase chain reaction, as described in the Methods section. Values were normalised to housekeeping genes (cyclophilin B and hypoxanthine guanine phosphoribosyltransferase), and are presented as mean ± SEM. Note the different scales in A and B, which better illustrates the differences in expression.
To determine the cellular localisation of Runx3 expression, immunohistochemistry was carried out. In the normal pancreas, Runx3 was present only in islet cells (fig 2A), with a predominantly cytoplasmic pattern, but was absent in normal ductal and acinar cells. In pancreatic cancer sections, Runx3 immunostaining was also seen in the islets of almost half of the samples (figure 2C, E). Interestingly, however, Runx3 expression was also evident in the cancer cells of seven of the 24 tumours analysed, and these tumours also displayed the highest Runx3 mRNA values by QRT-PCR. Although Runx3 expression was moderate in most of these tumours (fig 2B), some tumours displayed relatively strong Runx3 staining, which was seen in the cytoplasm and a considerable number of nuclei (fig 2C, D). Thirty seven of the 52 tumours showed weak to absent Runx3 staining (fig 2E, F). In three of the seven PDAC samples that exhibited Runx3 staining in the cancer cells, approximately 95% of the cancer cells were Runx3 positive, whereas in the other four samples, approximately 30–40% of the cancer cells exhibited Runx3 immunoreactivity. There was no obvious preferential localisation of the Runx3 positive tumours cells (for example, tumour periphery or centre). In addition, metastatic tumours were completely devoid of or showed only very weak Runx3 immunoreactivity (fig 3C, D). Interestingly, Runx3 immunoreactivity was present in the lymphocytes of six of the 16 samples that had a pronounced infiltration of lymphocytes (figs 2B, C and 3B). Runx3 positive lymphocytes were also present in CP-like lesions in PDAC samples. Interestingly, tubular complexes in these CP-like lesions adjacent to the tumour also showed Runx3 immunoreactivity in all but one case (data not shown). There was only scarce lymphocytic infiltration in the normal pancreas, and these lymphocytes were devoid of Runx3 immunoreactivity.
Figure 2.
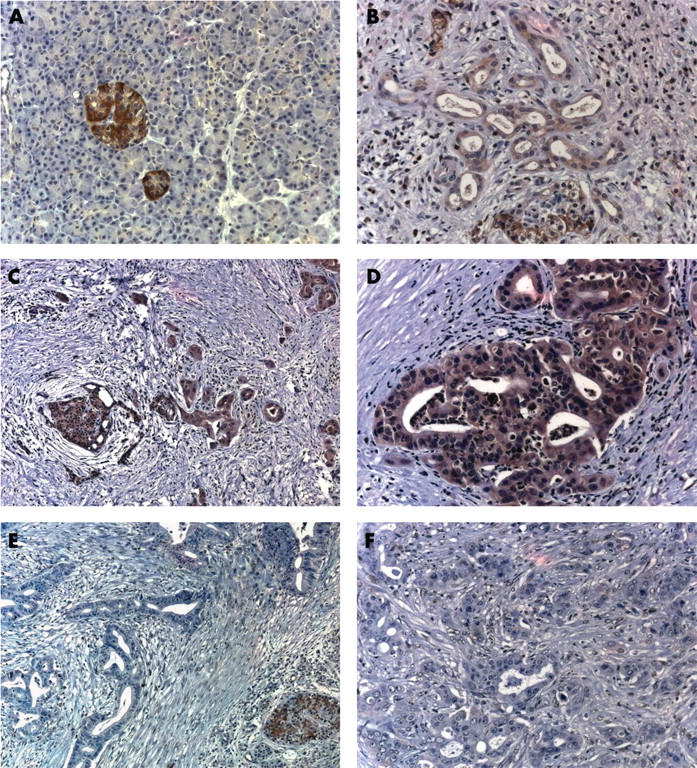
Runx3 localisation in pancreatic tissues: immunohistochemistry using a Runx3 specific antibody was carried out in (A) the normal pancreas and (B–F) pancreatic cancers. (B) A moderately stained pancreatic cancer with infiltrating inflammatory cells; (C, D) Runx3 positive cancers; (E, F) Runx3 negative pancreatic cancers.
Figure 3.
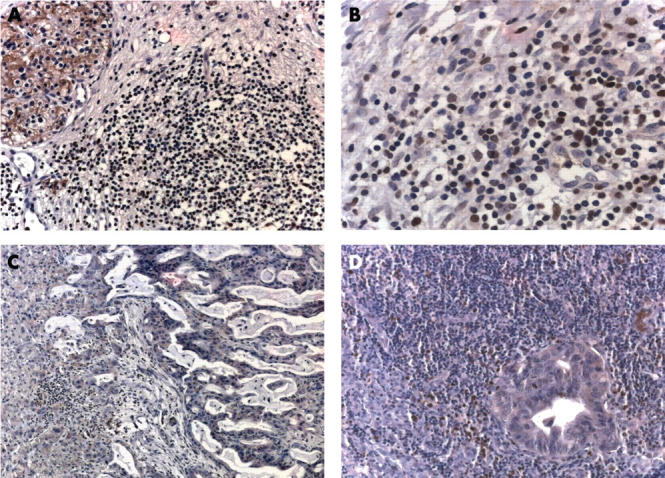
Runx3 localisation in primary and metastatic pancreatic cancer tissues: immunohistochemistry of Runx3 in (A and B) primary pancreatic cancers, (C) liver metastasis, and (D) lymph node metastasis. (A) Runx3 negative lymphocytes; (B) Runx3 immunoreactivity in some of the lymphocytes. (D) Note also some Runx3 positive lymphocytes surrounding the lymph node metastasis.
Runx3 expression and regulation in cultured pancreatic cancer cell lines
By QRT-PCR analysis, Runx3 expression was readily apparent in Colo-357 and T3M4 cells, low in Aspc-1, MiaPaCa-2, and Panc-1 cells, and below the level of detection in BxPc-3 and Capan-1 cells. Western blotting was used to evaluate Runx3 protein expression in pancreatic cancer cell lines. Runx3 was expressed in various amounts in the pancreatic cancer cell lines tested, with the highest expression seen in Colo-357 and T3M4 cells (fig 4A), in partial agreement with the QRT-PCR results. Interestingly, two immunoreactive bands migrating slightly above 40 kDa were present in some cell lines (BcPc-3, Colo-357, and T3M4), whereas in the other cell lines only the upper of these two bands could be detected. This is partially in agreement with previous reports showing that Runx3 yields two protein isoforms of 45 and 47.5 kDa by Western blotting.13
Figure 4.
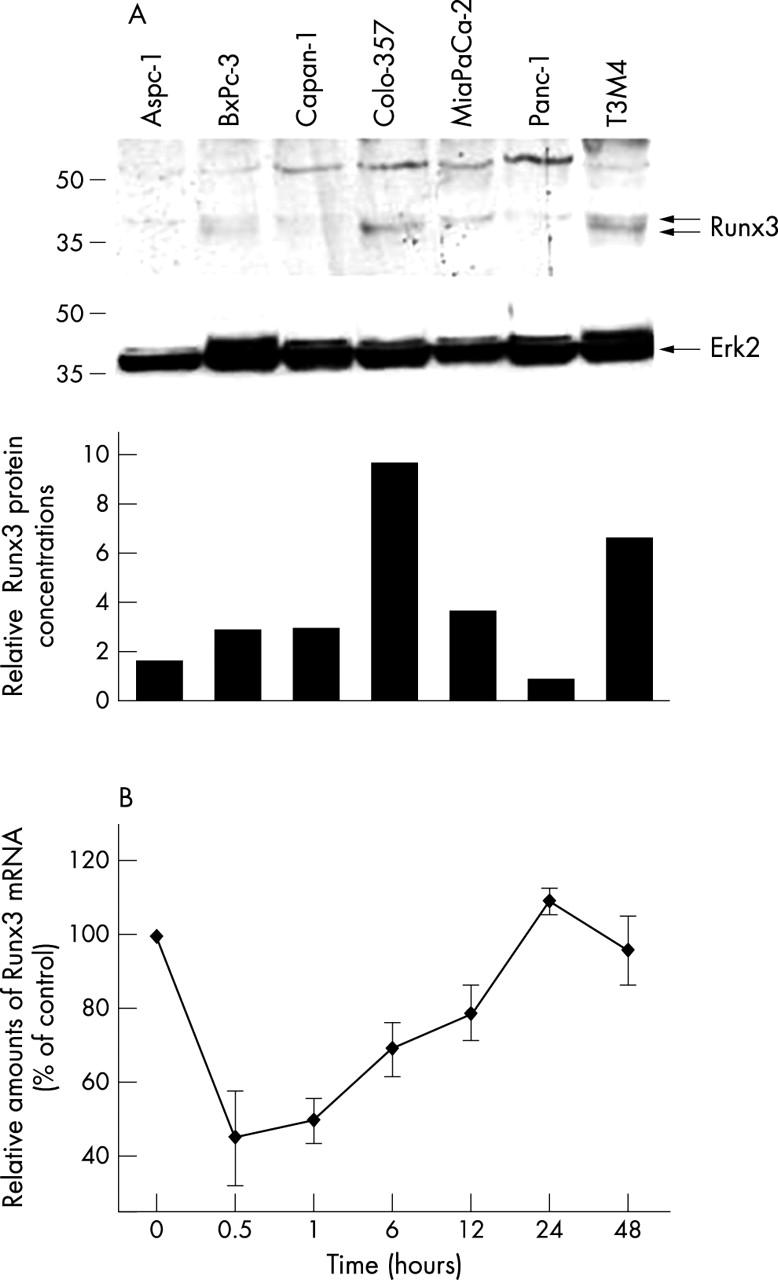
Runx3 expression and regulation in pancreatic cancer cell lines: (A) protein lysates (20 µg) of the indicated cell lines were subjected to Western blotting using a Runx3 antibody, as described in the Methods section (upper panel). Equal loading was determined by stripping the membrane and blotting with an antibody to Erk2 (middle panel). Relative amounts of Runx3 protein were determined by OD (Runx3)/OD (Erk2) × 100 for each sample (lower panel). (B) Colo-357 cells were incubated in the absence (0) or presence of 200pM transforming growth factor β1 for the indicated time. RNA was extracted and real time quantitative polymerase chain reaction for Runx3 was performed. The respective control mRNA expression values were set to 100%. Results are presented as mean ± SEM of three independent experiments. OD, optical density.
To investigate a possible transcriptional regulation of Runx3 by TGFβ1 in cultured pancreatic cancer cell lines, cells were treated with TGFβ1 (200pM) for up to 48 hours. After TGFβ1 treatment, Runx3 expression was repressed in Colo-357 for 12–24 hours and returned to pretreatment values thereafter (fig 4B). In contrast, in the other cell lines TGFβ1 did not influence Runx3 mRNA values (data not shown).
DISCUSSION
The pathogenesis of PDAC involves genetic alterations, such as K-ras protooncogene mutations, mutations of the p53, p16, and Smad4 tumour suppressor genes, and other less common mutations.2 In addition, there are numerous epigenetic alterations, including altered expression of several growth factors and their receptors.3 For example, PDACs overexpress all TGFβ isoforms and their receptors, and overexpression of these ligands and receptors is often associated with shortened postoperative survival of patients with pancreatic cancer.14,15
The TGFβ pathway is carefully regulated, with Smad proteins as the key component in the signal transduction pathway. In addition, other regulators, such as transcription factors, which facilitate Smad binding to target promoters, may provide routes for feedback and crosstalk.16 For example, members of the CBFA (core binding factor A) family of transcription factors act both as targets and partners of activated Smads. This family, also termed the “Runx family”, includes three human members: Runx1 (CBFA2/AML1), Runx2 (CBFA1/AML3), and Runx3 (CBFA2/AML2).17 These genes share a high degree of sequence similarity within most of their coding regions, yet they mediate distinct biological signals, and play pivotal roles during normal development and in neoplasia. Runx3 is a neurospecific transcription factor. Thus, in its absence, neurones in dorsal root ganglia do not survive long enough to extend their axons towards target cells, resulting in a lack of connectivity and ataxia.8 Runx3 is also highly expressed in thymic medulla and cortex, and functions in the lineage specification and homeostasis of CD8 positive T cells.18 Thus, Runx3 deficient cytotoxic T cells have defective responses to antigens9 and show impaired CD8 T cell maturation in the thymus.18 It has recently been shown that Runx3 is a tumour suppressor gene in gastric cancer. Gastric mucosal hyperplasia has been seen in Runx3 knockout mice, and the degree of loss of Runx3 expression correlated with the extent of the disease.7 It was also suggested that in Runx3 deficient mice, TGFβ1 induced cell death was strongly impaired, and therefore that Runx3 is a target of the TGFβ signalling pathway in vivo.7
In our present study, Runx3 expression was found only in islets in normal pancreatic tissues. In contrast, in pancreatic cancer tissues, Runx3 was present in both islets and cancer cells. Of interest, Runx3 expression was seen mainly in the cytoplasm of the cancer cells, although nuclear staining was seen in a considerable number of tumour cells. It is currently not known whether the observed cytoplasmic location of Runx3 has any functional consequences. Nonetheless, these observations differ from the findings in gastric tissues, in which Runx3 expression was lost in 45–60% of gastric cancer cells, but present in normal gastric epithelial cells.7 These results suggest that Runx3 does not act as a tumour suppressor in PDAC, because it was not expressed in normal ductal cells, but was expressed in some pancreatic ductal adenocarcinomas. Interestingly, all metastases were devoid of or displayed only very faint Runx3 immunoreactivity. It is also of interest that Runx3 mRNA values were high in CP, a known preneoplastic condition, suggesting that Runx3 expression is increased early in the pathogenesis of pancreatic cancer and lost at later steps in many pancreatic tumours. It is currently not known whether the Runx3 expression seen in some pancreatic tumours has a distinct pathogenetic role, and further studies are necessary to investigate this point. It is worthwhile to mention, however, that some groups have advocated islet cells as the cells of origin of PDAC.19 Thus, it could be hypothesised that there is indeed loss of Runx3 expression in some primary tumours compared with normal islets, and almost complete loss in metastatic tumours, pointing to a tumour suppressor role for Runx3 in pancreatic cancer.
“Runx3 mRNA values were high in chronic pancreatitis, a known preneoplastic condition, suggesting that Runx3 expression is increased early in the pathogenesis of pancreatic cancer and lost at later steps in many pancreatic tumours”
The regulation of Runx3 expression in pancreatic cancer is currently unknown. In our experiments, TGFβ1 repressed Runx3 expression only in Colo-357 pancreatic cancer cells, which have a functional TGFβ pathway.20 Interestingly, TGFβ1 did not alter Runx3 expression in Panc-1 pancreatic cancer cells with a functional TGFβ pathway, or in other cell lines that exhibit alterations, such as Smad4 mutations or low expression of the TGFβ receptor I.21 In addition, in BxPc-3 pancreatic cancer cells, which harbour a Smad4 deletion but that are nonetheless growth inhibited by TGFβ1,22 this growth factor did not alter Runx3 mRNA values. Because in vivo PDACs often exhibit alterations of the TGFβ signalling pathway,14,15,23–25 it is possible that the repressive effect of TGFβ on Runx3 expression is lost in tumours with alterations in this pathway, leading to the observed upregulation of Runx3.
Interestingly, in some pancreatic cancer samples, strong nuclear Runx3 expression was also seen in lymphocytes. This is consistent with previous reports of Runx3 expression in thymic medulla, spleen, and bone marrow,18 in addition to cells of haemopoietic origin, such as B cell lymphoma cells, myelomonoblastic leukaemia cells, and early myoblast cells.26 Runx3 was reported to have a crucial function in the lineage specification, maturation, and homeostasis of CD8 positive T cells.9 It has also been reported that Runx3 is induced by TGFβ1 in splenic B cells in the presence of activation signals,13 and that Runx3, in addition to Smad3/4 and cAMP response element binding protein, binds independently to the respective DNA sequences in the immunoglobulin α promoter, resulting in maximal TGFβ induced transcription and class switching to IgA.27 Because TGFβs are frequently overexpressed in PDAC,14 it is possible that cancer derived TGFβs induce Runx3 expression in infiltrating lymphocytes. TGFβs are strong immunosuppressors that are involved in the immunological escape of pancreatic cancer cells. However, the functional consequence of TGFβ induced Runx3 expression in tumour infiltrating lymphocytes is currently not known.
Take home messages.
In the normal pancreas, Runx3 is expressed only in the islets, whereas a considerable proportion of pancreatic tumour cells express Runx3
In pancreatic tumours Runx3 expression is localised in the tumour cells and the infiltrating lymphocytes
In cultures pancreatic tumour cell lines Runx3 expression can be repressed by transforming growth factor β1 (TGFβ1)
Runx3 might play a role in the pathogenesis of pancreatic cancer and there may be a connection with the frequently altered TGFβ pathway in this disease
In conclusion, in the normal pancreas, Runx3 expression is restricted to islet cells, whereas in PDAC it is also expressed in the cancer cells themselves and in infiltrating lymphocytes. Furthermore, its expression can be repressed by TGFβ1 in responsive pancreatic cancer cells. Together, these results suggest that Runx3 might play a role in the pathophysiology of PDAC, which is distinct from its role in gastric cancer, and that there is a connection with the frequently altered TGFβ pathway in this disease.
Acknowledgments
We thank Y Groner (Department of Molecular Genetics, The Weizmann Institute of Science, Rehovot, Israel) for providing us with the anti-Runx3 antibody, and for carefully proofreading the manuscript.
Abbreviations
CP, chronic pancreatitis
QRT-PCR, real time quantitative polymerase chain reaction
SDS, sodium dodecyl sulfate
TBS-T, Tris buffered saline-Tween
TGFβ, transforming growth factor β
REFERENCES
- 1.Jemal A, Thomas A, Murray T, et al. Cancer statistics, 2002. CA Cancer J Clin 2002;52:23–47. [DOI] [PubMed] [Google Scholar]
- 2.Kern SE, Hruban RH, Hidalgo M, et al. An introduction to pancreatic adenocarcinoma genetics, pathology and therapy. Cancer Biol Ther 2002;1:607–13. [DOI] [PubMed] [Google Scholar]
- 3.Ozawa F, Friess H, Tempia-Caliera A, et al. Growth factors and their receptors in pancreatic cancer. Teratog Carcinog Mutagen 2001;21:27–44. [DOI] [PubMed] [Google Scholar]
- 4.Karsenty G. Role of Cbfa1 in osteoblast differentiation and function. Semin Cell Dev Biol 2000;11:343–6. [DOI] [PubMed] [Google Scholar]
- 5.Tracey WD, Speck NA. Potential roles for RUNX1 and its orthologs in determining hematopoietic cell fate. Semin Cell Dev Biol 2000;11:337–42. [DOI] [PubMed] [Google Scholar]
- 6.Westendorf JJ, Hiebert SW. Mammalian runt-domain proteins and their roles in hematopoiesis, osteogenesis, and leukemia. J Cell Biochem 1999;32–33(suppl):51–8. [DOI] [PubMed] [Google Scholar]
- 7.Li QL, Ito K, Sakakura C, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 2002;109:113–24. [DOI] [PubMed] [Google Scholar]
- 8.Levanon D, Bettoun D, Harris–Cerruti C, et al. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J 2002;21:3454–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taniuchi I, Osato M, Egawa T, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 2002;111:621–33. [DOI] [PubMed] [Google Scholar]
- 10.Weinstein M, Yang X, Deng C. Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev 2000;11:49–58. [DOI] [PubMed] [Google Scholar]
- 11.Giese NA, Raykov Z, DeMartino L, et al. Suppression of metastatic hemangiosarcoma by a parvovirus MVMp vector transducing the IP-10 chemokine into immunocompetent mice. Cancer Gene Ther 2002;9:432–42. [DOI] [PubMed] [Google Scholar]
- 12.Levanon D, Brenner O, Negreanu V, et al. Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech Dev 2001;109:413–17. [DOI] [PubMed] [Google Scholar]
- 13.Shi MJ, Stavnezer J. CBF alpha3 (AML2) is induced by TGF-beta1 to bind and activate the mouse germline Ig alpha promoter. J Immunol 1998;161:6751–60. [PubMed] [Google Scholar]
- 14.Friess H, Yamanaka Y, Buchler M, et al. Enhanced expression of transforming growth factor beta isoforms in pancreatic cancer correlates with decreased survival. Gastroenterology 1993;105:1846–56. [DOI] [PubMed] [Google Scholar]
- 15.Friess H, Yamanaka Y, Buchler M, et al. Enhanced expression of the type II transforming growth factor beta receptor in human pancreatic cancer cells without alteration of type III receptor expression. Cancer Res 1993;53:2704–7. [PubMed] [Google Scholar]
- 16.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol 2000;1:169–78. [DOI] [PubMed] [Google Scholar]
- 17.Otto F, Lubbert M, Stock M. Upstream and downstream targets of RUNX proteins. J Cell Biochem 2003;89:9–18. [DOI] [PubMed] [Google Scholar]
- 18.Woolf E, Xiao C, Fainaru O, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A 2003;100:7731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pour PM, Pandey KK, Batra SK. What is the origin of pancreatic adenocarcinoma? Mol Cancer 2003;2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleeff J, Korc M. Up-regulation of transforming growth factor (TGF)-beta receptors by TGF-beta1 in COLO-357 cells. J Biol Chem 1998;273:7495–500. [DOI] [PubMed] [Google Scholar]
- 21.Wagner M, Kleeff J, Lopez ME, et al. Transfection of the type I TGF-beta receptor restores TGF-beta responsiveness in pancreatic cancer. Int J Cancer 1998;78:255–60. [DOI] [PubMed] [Google Scholar]
- 22.Kleeff J, Wildi S, Friess H, et al. Ligand induced upregulation of the type II transforming growth factor (TGF-beta) receptor enhances TGF-beta responsiveness in COLO-357 cells. Pancreas 1999;18:364–70. [DOI] [PubMed] [Google Scholar]
- 23.Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 1996;271:350–3. [DOI] [PubMed] [Google Scholar]
- 24.Kleeff J, Maruyama H, Friess H, et al. Smad6 suppresses TGF-beta-induced growth inhibition in COLO-357 pancreatic cancer cells and is overexpressed in pancreatic cancer. Biochem Biophys Res Commun 1999;255:268–73. [DOI] [PubMed] [Google Scholar]
- 25.Kleeff J, Ishiwata T, Maruyama H, et al. The TGF-beta signaling inhibitor Smad7 enhances tumorigenicity in pancreatic cancer. Oncogene 1999;18:5363–72. [DOI] [PubMed] [Google Scholar]
- 26.Le XF, Groner Y, Kornblau SM, et al. Regulation of AML2/CBFA3 in hematopoietic cells through the retinoic acid receptor alpha-dependent signaling pathway. J Biol Chem 1999;274:21651–8. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Derynck R. Transcriptional regulation of the transforming growth factor-beta-inducible mouse germ line Ig alpha constant region gene by functional cooperation of Smad, CREB, and AML family members. J Biol Chem 2000;275:16979–85. [DOI] [PubMed] [Google Scholar]


