Abstract
Aims: To characterise a novel strain of adenovirus (Ad) type Ad8 (genome type Ad8I) involved in an epidemic keratoconjunctivitis (EKC) outbreak in Hiroshima city using serological testing and sequence analysis of the fibre and hexon gene.
Methods: A neutralisation test (NT) was performed in microtitre plates containing a confluent monolayer of A549 cells using 100 tissue culture infectious doses of virus and type specific antisera. The haemagglutination inhibition test was also carried out in microtitre plates with rat erythrocytes using four haemagglutination units of virus and twofold dilutions of serum. The fibre gene was sequenced by generating overlapping polymerase chain reaction products or by direct sequencing of genomic DNA. Primer selection was based on alignment of the fibre genes of human adenovirus serotypes Ad8, Ad19, Ad37, Ad9, and Ad15 available from Gene Bank.
Results: The virus strain was specifically neutralised by anti-Ad8 antibodies, although there was a major crossreaction with anti-Ad9 antibodies. Haemagglutination was equally inhibited by anti-Ad8 and anti-Ad9 antibodies. The predicted amino acid sequences of the hypervariable regions (HVRs) of the Ad8I hexon gene showed higher homology with Ad9 (83.3%) than with Ad8 (62.0%). However, the Ad8I fibre knob was more homologous to Ad8 (94.4%) than to Ad9 (91.6%).
Conclusions: Ad8I is a unique strain of adenovirus because of its lower genomic homology with Ad8, major crossreactivity with Ad9 in NT, and mixed genetic organisation of HVRs of the hexon gene. These factors may have enabled the virus to circumvent acquired immunity, resulting in the outbreak.
Keywords: adenovirus, epidemic keratoconjunctivitis, hexon gene, fibre gene, sequence analysis
There are 51 serotypes of human adenoviruses (Ads), which are divided into six subgenera (A–F) based on biochemical, immunological, and morphological criteria.1,2 Among them, subgenus D consists of 32 serotypes including Ad8, Ad19, and Ad37, the main agents of epidemic keratoconjunctivitis (EKC), and Ad9 and Ad15, which cause acute follicular conjunctivitis.3,4 EKC is a clinical disease entity characterised by severe bilateral conjunctivitis, with substantial corneal and extraocular involvement, leading to decreased quality of life and possible economic consequences.
Between 1995 and 1997, in Hiroshima city, a novel strain of Ad8 was isolated from an outbreak of EKC and sporadic cases of EKC; this strain was identified as genome type Ad8I by restriction endonuclease analysis (REA). Before the detection of Ad8I, genome types A8A–H had been identified by REA.5 The Ad8 genome types usually share 96% and > 90% homology when compared by REA or the predicted amino acid sequence of the hypervariable regions (HVRs) of the hexon gene/protein, respectively.5 In contrast, the novel strain Ad8I was only 71% and 62.0% homologous to other Ad8 genome types when REA and sequence analysis of the HVRs of the hexon gene were carried out. This unusually low degree of homology characterised Ad8I as a unique strain among the Ad8 genome types described so far, prompting us to study this strain in depth at the molecular level.
“Epidemic keratoconjunctivitis is a clinical disease entity characterised by severe bilateral conjunctivitis, with substantial corneal and extraocular involvement, leading to decreased quality of life and possible economic consequences”
The hexon protein of adenovirus carries type specific antigenic determinants in its HVRs, which react with neutralising antibody in the serum neutralisation test (NT). Neutralisation of the infectivity of adenoviruses is primarily determined by antibody against the hexon protein. In contrast, the fibre protein is responsible for haemagglutination and also the attachment of the virus to specific cellular receptors through its receptor binding site on the knob region. Together with the hexon protein it is responsible for the serotype specificity of adenoviruses.6–9 In our present study, the fibre and hexon genes of the novel strain Ad8I were analysed to compare immunological data (NT and haemagglutination inhibition (HAI) test) with the molecular biology results in an attempt to define any genetic differences that might be relevant to the outbreak of EKC.
MATERIALS AND METHODS
Viruses
Adenovirus prototypes Ad9 and Ad11 and the antisera were obtained from American Type Culture Collection (ATCC, Rockville, Maryland, USA); Ad8 was obtained from the collection of the National Institute of Infectious Diseases, Tokyo, Japan. The Ad8I strain was isolated in Hiroshima city, Japan, from the outbreak and sporadic cases of EKC from 1995 to 1997.
Serological tests
For NT analysis the adenovirus stock was grown in A549 cells and titres were determined in microtitre plates containing a confluent monolayer of A549 cells. Aliquots of 25 µl of 100 tissue culture infectious doses of virus (100TCID50) were incubated at 37°C for 60 minutes with 25 µl volumes of twofold serially diluted rabbit antiserum, and inoculated into the A549 cells. The adenovirus type was determined by the antiserum, which completely inhibited viral growth after 72 hours of incubation. The HAI test was also carried out in microtitre plates with rat erythrocytes. Briefly, four haemagglutination units of virus and twofold dilutions of serum (which had been treated at 65°C for 30 minutes to remove non-specific agglutinin), both in a 25 µl volume, were incubated at room temperature for one hour. Next, 25 µl (final concentration, 1.0%) of erythrocytes was added and the mixture was allowed to stand for one hour at 37°C, after which the results were read. The antisera to Ad8, Ad9, Ad19, and Ad11 in both the tests were obtained from ATCC. The rat blood cells were obtained from Cosmobio (Tokyo, Japan).
Extraction of viral DNA
Viral DNA was extracted from a confluent monolayer of Hep2 cells inoculated with virus stock. When a cytopathic effect of greater than 75% was observed, cells were dislodged and pelleted by low speed centrifugation. Cells were washed in phosphate buffered saline and resuspended in lysis buffer (10mM Tris/HCl (pH 7.4), 10mM EDTA, 1% sodium dodecyl sulfate) for 15 minutes at room temperature. The suspension was incubated with proteinase K (Sigma Chemical, St Louis, Missouri, USA) at a concentration of 200 µg/ml at 37°C for one hour. Next, 5M NaCl was added to a final concentration of 1M, and further incubated at 4°C overnight to precipitate cellular DNA. The suspension was centrifuged at 15 000 ×g for 30 minutes. The supernatant was incubated with 30 µg of RNase A (Sigma Chemical) for one hour and then extracted twice in phenol/chloroform, after which the supernatant was precipitated in two volumes of absolute ethanol. After drying, the DNA was suspended in 50 µl of TE buffer (10mM Tris/HCl (pH 7.4), 10mM EDTA) and measured spectrophotometrically.
DNA sequencing
The fibre gene was sequenced by generating overlapping polymerase chain reaction (PCR) products or by direct cycle sequencing of genomic DNA. The selection of the primers was based on available sequences from Gene Bank (Gene Bank accession numbers for the fibre gene: X74660 (Ad8), U69731 (Ad19a), U69132 (Ad37), X74659 (Ad9), and X72934 (Ad15)) of human adenovirus serotypes Ad8, Ad19, Ad37, Ad9, and Ad15. All products were sequenced from both directions with internal and template primers (table 1). Full length adenoviral DNA, extracted by the modified Hirt’s method, was used as a template for PCR. The PCR amplification was carried out in 50 µl reaction mixtures containing 1 μl of DNA, 5 µl of 10× concentrated buffer, 0.5 µM of each primer pair, 200 µM of each dNTP, and 1.25 U of Taq polymerase (Boehringer-Mannheim, Mannheim, Germany). The assays were performed in a programmable heat block (Perkin Elmer model 9600-R; Foster City, California, USA). Thermal cycling consisted of preliminary denaturation for three minutes at 94°C, followed by 35 cycles of denaturation at 94°C for one minute, annealing at 47°C for one minute, and extension at 72°C for two minutes, with a final extension at 72°C for seven minutes. The amplification products were analysed on a 1.5% agarose gel. The PCR products were purified using a DNA fragment purification kit (Mag Extractor-PCR and Gel Clean Up; Toyobo Co Ltd, Osaka, Japan), according to the manufacturer’s protocol. The cycle sequence reaction was carried out with an ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Chiba, Japan). The sequences were determined by a Genetic Analyser 310 (Applied Biosystem, Foster City, California, USA). The hexon gene sequence of Ad8I was previously deposited in DDBJ/Gene Bank under the accession number AB090344. The nucleotide sequences of both the hexon and fibre genes were translated into amino acid sequences and compared with the available sequences of Ad8, Ad9, Ad15, Ad19a, and Ad37 by DNASIS (Hitachi Software Ltd, Tokyo, Japan).
Table 1.
Oligonucleotide primers for polymerase chain reaction and sequencing of the adenovirus fibre gene
| Primer | Polarity | Position | Sequence (5′–3′) |
| AdnF1 | + | 1–17 | 5′-AAG GGA TGT CAA ATT CC-3′ |
| ShaftF | + | 274–298 | 5′-CTG GAA AAT TAA CAG TTA ATA CTG A-3′ |
| Knob1F | + | 513–530 | 5′-AGA GTT GGA GAA GGC GGC-3′ |
| Knob2F | + | 813–833 | 5′-TCT TCA AAT CTT GGT AAA TCA-3′ |
| ShaftR | − | 297–274 | 5′-CAG TAT TAA CTG TTA ATT TTC CAG-3′ |
| Knob1R | − | 530–513 | 5′-GCC GCC TTC TCC AAC TCT AA-3′ |
| Knob2R | − | 833–813 | 5′-TGA TTT ACC AAG ATT TGA AGA-3′ |
| AdDRcom | − | 1209–1188 | 5′-TAC YCG YGC TGG TGT AAA AAT C-3′ |
Nucleotide sequence accession numbers
The nucleotide sequence data reported here will appear in the DDBJ/Gene Bank nucleotide sequence database with the accession number AB098565 (fibre gene).
RESULTS
Serological tests
Ad8I crossreacted with Ad8 and Ad9 antisera in both the NT and the HAI test (table 2).
Table 2.
NT and HAI titres of the Ad8, Ad8I, and Ad9 strains
| Adenovirus | Ad8 | Ad8I | Ad9 |
| NT titration | |||
| Ad8 antiserum | 256 | 256 | 16 |
| Ad9 antiserum | <4 | 128 | 256 |
| HAI titration | |||
| Ad8 antiserum | 320 | 320 | 320 |
| Ad9 antiserum | 320 | 320 | 320 |
| Ad19 antiserum | 20 | 20 | 20 |
| Ad11 antiseum | <5 | <5 | <5 |
Ad, adenovirus; HAI, haemagglutination inhibition; NT, neutralisation test.
Sequence analysis
Fibre gene: nucleotide sequence
We determined the nucleotide sequence of the Ad8I fibre gene. Sequence analysis showed that the fibre region of this strain is 1083 nucleotides long, whereas corresponding regions of the fibre genes of Ad8 and Ad9 are 1086 nucleotides long. The difference in length results from the deletion of three nucleotides in the shaft region. Ad8I shows higher homology with Ad8 (overall homology, 95.5%) than with Ad9 (overall homology, 91.5%). However, in the knob region it shows 97.0% and 92.7% homology with Ad8 and Ad9, respectively.
Fibre polypeptide
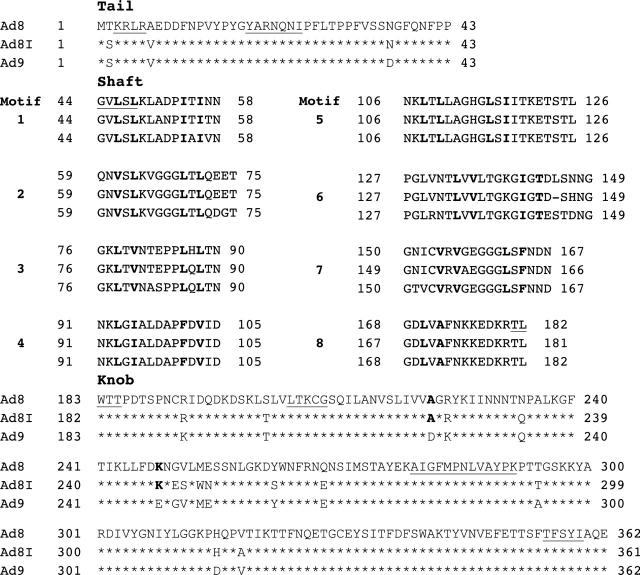
The amino acid sequence of the Ad8I fibre gene was deduced from the nucleotide sequences and aligned for maximum homology (fig 1). The predicted fibre polypeptide comprises 361 amino acid residues, whereas the fibre polypeptides of Ad8 and Ad9 comprise 362 amino acids each. The shaft of Ad8I is 138 residues long, whereas the corresponding regions of Ad8 and Ad9 are made up of 139 amino acids. It also has eight repeats of a 15 amino acid basic structural unit (motif), containing conserved hydrophobic glycine (G) and proline (P) residues. These eight motif repeats in the shaft are consistent with the length of the subgenus D fibre protein. Ad8I showed six mismatches with Ad8 and 16 mismatches with Ad9, resulting in 90.5% and 84.0% homology, respectively.
Figure 1.
Comparison of predicted amino acid sequences of Ad8, Ad8I, and Ad9 fibre polypeptides. The structural domains tail, knob, and shaft are shown, and the eight repeating shaft motifs are numbered. The sequences are aligned to obtain maximal homology. Amino acids identical to Ad8 in the tail and knob are marked by asterisks. The conserved hydrophobic residues are in bold type. Sequences conserved among the subgenus D adenoviruses are underlined.
The fibre knob of Ad8I comprises 180 amino acids—the same as Ad8 and Ad9—and shows nine and 12 residue mismatches with Ad8 and Ad9, resulting in 94.4% and 91.6% homology, respectively (table 3). In the knob, residues are conserved between Ad8 and Ad8I, including alanine (A) in position 222 and lysine (K) in position 247, which are replaced by aspartic acid (D) and glutamic acid (E), respectively, in Ad9 (fig 1).
Table 3.
Homology (%) between fibre nucleotide and amino acid sequences of Ad8I and those of the members of the adenovirus subgenus D
| Fibre region | Ad8 | Ad9 | Ad15 | Ad19a/37 |
| Tail | ||||
| DNA | 95.4 | 96.8 | 95.3 | 96.1 |
| Protein | 93.0 | 97.6 | 95.3 | 95.3 |
| Shaft | ||||
| DNA | 94.0 | 88.1 | 59.1 | 53.8 |
| Protein | 90.5 | 84.0 | 49.2 | 31.8 |
| Knob | ||||
| DNA | 97.0 | 92.7 | 54.2 | 78.9 |
| Protein | 94.4 | 91.6 | 35.5 | 77.6 |
| Overall | ||||
| DNA | 95.5 | 91.5 | 61.0 | 71.1 |
| Protein | 92.7 | 89.4 | 47.9 | 62.0 |
DNA and amino acid homologies were obtained using DNASIS (Hitachi) software.
Hexon gene
The amino acid sequences of the HVRs of the Ad8I hexon protein showed 83.3% homology with Ad9, whereas only 62.0%. 70.9%, and 63.6% homology was seen with Ad8, Ad19, and Ad37, respectively (table 4). Ad8I did not show the unique insertion of 33 residues in HVR1 as is seen in Ad8 (fig 2).
Table 4.
Hexon gene amino acid homologies (%) of Ad8I with the members of adenovirus subgenus D
| Ad | HVR1 | HVR2 | HVR3 | HVR4 | HVR5 | HVR6 | HVR6 | Overall |
| Ad8 | 8.8 | 11.7 | 18.1 | 52.3 | 10.5 | 41.6 | 42.8 | 62.0 |
| Ad9 | 61.7 | 94.1 | 81.8 | 23.8 | 71.4 | 41.6 | 77.2 | 83.3 |
| Ad15 | 45.4 | 29.4 | 27.2 | 26.3 | 33.3 | 58.3 | 75.0 | 75.0 |
| Ad19 | 41.1 | 11.7 | 45.4 | 38.0 | 33.3 | 41.6 | 65.9 | 70.9 |
| Ad37 | 14.7 | 41.1 | 63.6 | 20.0 | 14.2 | 58.3 | 29.5 | 63.6 |
Amino acid homologies were obtained using DNASIS (Hitachi) software.
Ad, adenovirus; HVR, hypervariable region.
Figure 2.
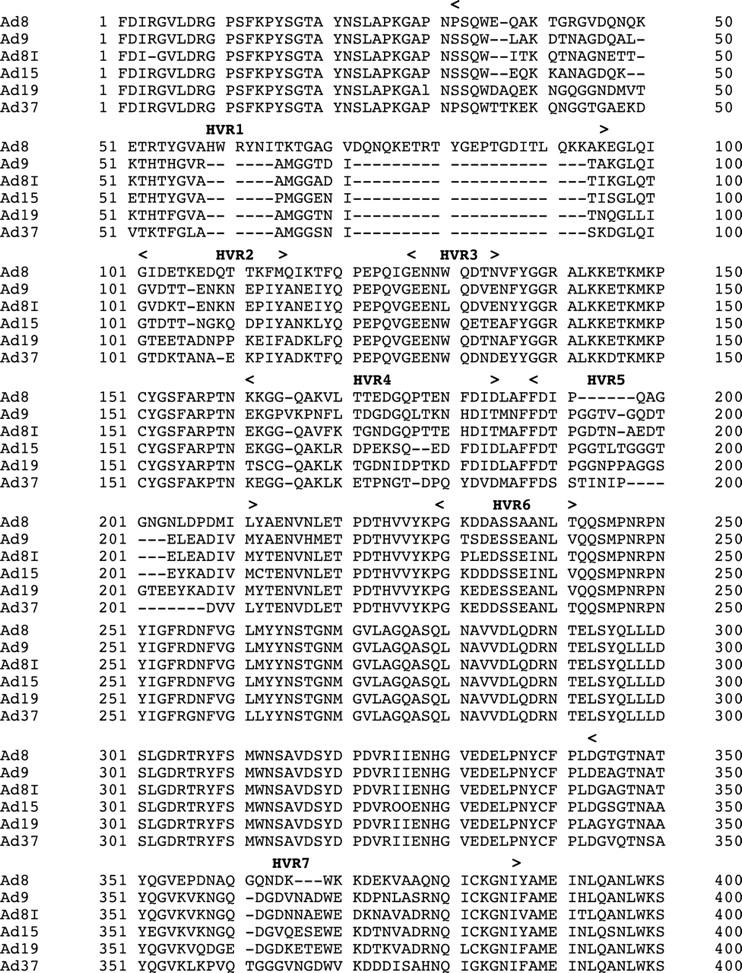
Comparison of predicted amino acid sequences of seven hypervariable regions (HVRs) of adenovirus 8 (Ad8), Ad9, Ad8I, Ad15, Ad19, and Ad37 hexon polypeptide. The sequences of loop 1 and loop 2 have been aligned to obtain maximal homology. The HVRs are marked above.
DISCUSSION
Adenoviral keratoconjunctivitis is endemic in Japan, with a peak incidence in the summer. According to the National Surveillance System of Infectious Diseases in Japan, adenovirus alone is responsible for 95% of viral conjunctivitis, with a predominance of subgenus D.10 In contrast to Ad19 and Ad37, Ad8 possesses a higher tropism for the conjunctiva and induces severe clinical manifestations in EKC, whereas Ad9 infrequently causes acute follicular conjunctivitis and mild respiratory infections.5,11 Although REA of the genome is useful for following the epidemiological distribution of the EKC strains, it cannot explain the tropism and infectivity of the virus at the molecular level. In a previous study, we investigated the circulation of the Ad8 genome type in a microenvironment over a prolonged period, and recovered the novel Ad8I genome type from an outbreak of EKC.5 In our subsequent study, we have shown that the HVRs of the Ad8I hexon are more homologous to Ad9 than to Ad8. To date, Ad9 has not been reported as an ocular pathogen in Japan. Therefore, the NT was re-performed with Ad9 antiserum using the Ad9 prototype as a control; Ad8I was neutralised by the anti-Ad8 antibody, although there was major crossreactivity with anti-Ad9.
x Ray crystallography of the human adenovirus hexon protein reveals that the hexon top consists of three long loops L1, L2, and L4. Sequence comparisons of several serotypes have shown that the seven hypervariable regions (HVRs) are located on L1 and L2, and form the virion surface. Two or more of these seven HVRs carry neutralisation epitopes that react with neutralising antibody in the NT.12 The L1 and L2 regions were found to be highly conserved within a serotype. Furthermore, 80% or more amino acid homology in the L1 and L2 portions of the hexon protein is thought to provide a basis of crossneutralisation among the serotypes (for example, Ad4 and Ad16; Ad3 and Ad7).13 In our study, the HVRs of Ad8I show 83.3% and 62.0% overall amino acid homology with Ad9 and Ad8, respectively. Despite such a low homology, there are two possible reasons for neutralisation by the anti-Ad8 antibody. First, the neutralisation epitopes in the hexon are complex and conformational rather than linear, and are composed of serotype specific residues, although the number and exact position of these residues in the HVRs are unknown. Therefore, it is possible that the recombination and mutational events that have taken place spared the neutralisation epitopes. Second, an antigenic determinant located in the fibre knob may also play a role in neutralisation.6
“We have shown that the hypervariable regions of the Ad8I hexon are more homologous to Ad9 than to Ad8”
In the HAI test, haemagglutination was equally inhibited by antisera against Ad8 and Ad9, a typical characteristic of Ad8 and Ad9 in this test.14 Comparison of the predicted amino acid homology of the isolate with the prototypes, in addition to the presence of specific residues in the fibre knob, apparently provided a guideline to distinguish EKC (Ad8) from non-EKC (Ad9) strain.15 The fibre knob amino acid sequence of the new strain shares 94.4% and 91.6% homology with Ad8 and Ad9, respectively. Seven conserved sequences (fig 1) found in the subgenus D fibre knob are also present in Ad8I. The new strain carries residues A (alanine) and K (lysine) in positions 222 and 247, which are conserved among EKC strains Ad8, Ad19, and Ad37, but are replaced by other residues in Ad9; these residues are thought to play an important role in the higher tropism of these strains for corneal and conjunctival cells.15 Hence, it seems likely that the Ad8I strain has the same Ad8 fibre ancestor, but has been modified slightly.
We have shown that the outbreak by the novel strain Ad8I can be explained by the immunological data and the molecular biology of the hexon and the fibre genes. The high homology between Ad8I and Ad8 in the fibre knob possibly played a key role in the higher tropism of this strain for conjunctival cells. In contrast, illegitimate recombination helped the virus to escape the acquired immune response present in the population and caused the outbreak of EKC.16 In conclusion, Ad8I is a unique strain because of its lower genomic homology with other Ad8 genome types, major crossreactivity with Ad9 in the NT, and unique genetic organisation at the HVRs of the hexon and the fibre genes, which possibly enhanced the pathogenic potential of the virus. It is of utmost importance to accumulate gene sequence data on the HVRs of the hexon and the fibre genes of EKC causing adenoviruses to help predict future outbreaks of adenovirus infections.
Take home messages.
Adenovirus 8I (Ad8I) is a unique strain of adenovirus because of its lower genomic homology with Ad8, major crossreactivity with Ad9 in the neutralisation test, and mixed genetic organisation of hypervariable regions of the hexon gene
The high homology between Ad8I and Ad8 in the fibre knob may explain the higher tropism of this strain for conjunctival cells
Illegitimate recombination in the hexon and fibre genes may have helped the virus to escape the acquired immune response present in the population, enabling this outbreak of epidemic keratoconjunctivitis to occur
Acknowledgments
Supported by grant-in-aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS), Japan.
Abbreviations
Ad, adenovirus
ATCC, American Type Culture Collection
EKC, epidemic keratoconjunctivitis
HAI, haemagglutination inhibition
HPV, hypervariable region
NT, neutralisation test
PCR, polymerase chain reaction
REA, restriction endonuclease analysis
REFERENCES
- 1.de Jong JC, Wermwnbol AG, Verweiji-Uijterwall MW, et al. Adenovirus from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotype Ad50 and Ad51 of species B1 and D, respectively. J Clin Microbiol 1999;37:3940–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shenk T. Adenoviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, eds. Fields virology, 5th ed. Philadelphia: Lippincort-Raven, 1996:2111–48.
- 3.Jawetz E, Kimura S, Nicholas AN, et al. New type of APC virus from keratoconjunctivitis. Science 1955;122:1190–2. [DOI] [PubMed] [Google Scholar]
- 4.Kemp MC, Hierholzer JC. The changing etiology of epidemic keratoconjunctivitis: antigenic and restriction enzyme analyses of adenovirus types 19 and 37 isolated over a 10-year period. J Infect Dis 1983;148:24–33. [DOI] [PubMed] [Google Scholar]
- 5.Adhikary AK, Numaga J, Kaburaki T, et al. Genetic characterization of adenovirus type 8 isolated in Hiroshima city over a 15 year period. J Clin Pathol 2003;56:120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adrian T, Wigand R, Wadell G. Serological and biochemical characteristics of intermediate adenovirus strains of subgenus D. Arch Virol 1987;97:347–57. [DOI] [PubMed] [Google Scholar]
- 7.Norby E. The structural and functional diversity of adenovirus capsid components. J Gen Virol 1969;5:221–36. [DOI] [PubMed] [Google Scholar]
- 8.Wadell G, Hammarskjold ML, Winberg G, et al. Genetic variability of adenoviruses. Ann N Y Acad Sci 1980;354:16–42. [DOI] [PubMed] [Google Scholar]
- 9.Valentine RC, Pereira HG. Antigen and structures of adenovirus. J Mol Biol 1965;13:13–20. [DOI] [PubMed] [Google Scholar]
- 10.Yamadera S, Yamashita K, Akatsuka, et al. Adenovirus surveillance, 1982–1995. Jpn J Med Sci Biol 1995;48:199–221. [PubMed] [Google Scholar]
- 11.Eiz B, Adrian T, Pring-Akerblom P. Immunological adenovirus variant strains of subgenus D: comparison of the hexon and fiber sequences. Virology 1995;213:313–20. [DOI] [PubMed] [Google Scholar]
- 12.Crawford-Miksza L, Schnurr David P. Adenovirus serotype evolution is driven by illegitimate recombination in the hypervariable regions of the hexon protein. Virology 1996;224:357–67. [DOI] [PubMed] [Google Scholar]
- 13.Li QG, Wadell G. Genetic variability of hexon loops 1 and 2 between seven genome types of adenovirus 7. Arch Virol 1999;144:1739–49. [DOI] [PubMed] [Google Scholar]
- 14.Hierholzer JC, Dowdle WR. Immunological basis of the adenovirus 8–9 cross-reaction. J Virol 1970;6:782–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pring-Akerblom P, Adrian T. Characterization of adenovirus subgenus D fiber genes. Virology 1995;206:564–71. [DOI] [PubMed] [Google Scholar]
- 16.Adhikary AK, Inada T, Suzuki E, et al. Characterisation of hexon and fibre genes of a novel strain of adenovirus involved in epidemic keratoconjunctivitis. J Clin Pathol 2004;57:95–7. [DOI] [PMC free article] [PubMed] [Google Scholar]