Abstract
Aim: The evaluation of allelic losses at the FHIT and the BRCA1 genes and at three other loci at the 17q region in a series of 34 sporadic breast cancer cases from Southern Brazil.
Methods: The samples were evaluated for loss of heterozygosity (LOH) at the FHIT and the BRCA1 genes and at three other microsatellite markers at 17q, and the findings were correlated with clinicopathological parameters.
Results: The BRCA1 intragenic marker, D17S855, had the highest frequency of LOH, detected in 10 of 24 informative cases, followed by the D17S579 (six of 23 informative cases), D17S806 (five of 21 informative cases), and D17S785 markers (five of 21 informative cases). LOH at the FHIT intragenic marker, D3S1300, was found in six of 25 informative cases. In four of the six cases with LOH of the FHIT gene, there was concomitant loss of the BRCA1 intragenic marker.
Conclusions: The frequency of allelic losses in the FHIT and BRCA1 loci in the Southern Brazilian population is similar to that described in the general population. No correlations were found when the total LOH frequency was compared with tumour size, grade, or presence of axillary lymph node metastasis. Further studies using larger sporadic breast cancer samples and additional markers would be useful to confirm these findings, in addition to establishing more specific associations with clinicopathological parameters in this specific population.
Keywords: loss of heterozygosity, sporadic breast cancer, BRCA1 gene, FHIT gene
Breast cancer is the most common malignancy in women and one in eight Brazilian women will probably develop breast cancer in their lifetime.1 Among the genetic alterations involved in the development and progression of breast cancer, allelic losses at particular chromosomal regions are common, and may indicate deletion of tumor suppressor genes.2
Loss of heterozygosity (LOH) at 17q has been reported in about 30–60% of sporadic breast cancer cases and in many studies involves the BRCA1 gene located at 17q21.3–5 However, in sporadic breast cancer, somatic mutations inactivating the BRCA1 gene are rare,6 and it has been suggested that epigenetic mechanisms of inactivation, in addition to LOH at this locus, may take place.5,7
“Among the genetic alterations involved in the development and progression of breast cancer, allelic losses at particular chromosomal regions are common, and may indicate deletion of tumor suppressor genes”
The region 3p14, where the FHIT gene is located, is a common site for chromosomal rearrangements in breast cancer,8 and contains the most active common fragile site in the human genome, the FRA3B locus.9 Genetic alterations with reduced expression of this gene are found in about 30% of breast cancers.10–12 LOH studies have shown allelic losses at 3p14 in about 25% of primary breast carcinomas, suggesting that FHIT may have suppressor-like properties.10,13 In familial breast cancer kindreds, early studies14 demonstrated a higher frequency of allelic imbalance at 3p14 compared with that seen in the sporadic breast cancers. More recent studies in BRCA2 positive tumours have shown a high frequency of FHIT LOH and reduced expression of the FHIT protein.12
In our study, we evaluated LOH at the BRCA1 and FHIT loci, in addition to three other loci on 17q, in a series of 34 sporadic breast cancer cases from Southern Brazil. The findings were analysed in relation to clinicopathological parameters.
MATERIALS AND METHODS
Samples
Primary tumour and peripheral blood specimens were obtained from 34 patients undergoing surgery for sporadic breast cancer at the Hospital Nossa Senhora das Graças and Hospital de Clinicas, Curitiba, Southern Brazil, with informed consent. The patients had no family history of breast cancer and the mean (SD) age was 54.4 (12.5) years. The tumours were classified as grade 2 (17 samples) and grade 3 (17 samples) invasive ductal carcinomas. Table 1 presents the clinicopathological information of the patients.
Table 1.
Clinicopathological information and LOH analysis results
| Sample | Histological grade | Tumour size (cm) | Age (years) | Lymph node metastasis | D17S855 | D17S579 | D17S806 | D17S579 | D3S1300 |
| CP 147 | IDC grade 2 | 4.0 | 35 | + | LOH | No LOH | No LOH | No LOH | NI |
| CP 149 | IDC grade 2 | 2.0 | 60 | + | No LOH | NI | No LOH | No LOH | NI |
| CP 158 | IDC grade 2 | 2.3 | 64 | + | NI | No LOH | NI | No LOH | No LOH |
| CP 161 | IDC grade 2 | 2.5 | 54 | + | No LOH | No LOH | No LOH | No LOH | NI |
| CP 165 | IDC grade 2 | 2.0 | 38 | + | No LOH | No LOH | No LOH | No LOH | No LOH |
| CP 169 | IDC grade 2 | 1.0 | 53 | Unknown | LOH | NI | No LOH | No LOH | No LOH |
| CP 176 | IDC grade 2 | 1.0 | 65 | − | LOH | NI | LOH | LOH | LOH |
| CP 213 | IDC grade 2 | 2.5 | 58 | − | NI | NI | No LOH | No LOH | NI |
| CP 227 | IDC grade 2 | 1.0 | 71 | Unknown | NI | No LOH | NI | NI | No LOH |
| CP 228 | IDC grade 2 | 1.2 | 44 | − | No LOH | NI | No LOH | No LOH | NI |
| CP 229 | IDC grade 2 | 2.0 | 78 | − | No LOH | NI | NI | No LOH | NI |
| CP 231 | IDC grade 2 | 2.0 | 54 | + | LOH | No LOH | NI | LOH | No LOH |
| CP 239 | IDC grade 2 | 3.2 | 48 | − | LOH | LOH | NI | NI | LOH |
| CP 253 | IDC grade 2 | 3.0 | 45 | + | NI | LOH | LOH | NI | No LOH |
| CP 255 | IDC grade 2 | 2.0 | 59 | + | NI | No LOH | No LOH | No LOH | No LOH |
| CP 257 | IDC grade 2 | 1.7 | 46 | − | No LOH | No LOH | No LOH | NI | No LOH |
| CP 271 | IDC grade 2 | 4.5 | 47 | + | No LOH | No LOH | No LOH | NI | No LOH |
| HC 102 | IDC grade 3 | 4.0 | 55 | + | NI | No LOH | NI | NI | No LOH |
| HC 146 | IDC grade 3 | 2.2 | 44 | Unknown | NI | LOH | NI | LOH | NI |
| HC 167 | IDC grade 3 | 3.0 | 45 | + | LOH | No LOH | NI | No LOH | LOH |
| CP 010 | IDC grade 3 | 2.0 | 65 | + | No LOH | LOH | No LOH | No LOH | No LOH |
| CP 160 | IDC grade 3 | 2.8 | 54 | − | No LOH | No LOH | NI | NI | No LOH |
| CP 183 | IDC grade 3 | 1.5 | 44 | Unknown | NI | NI | No LOH | NI | No LOH |
| CP 212 | IDC grade 3 | 3.5 | 75 | + | No LOH | NI | NI | No LOH | No LOH |
| CP 224 | IDC grade 3 | 4.0 | 76 | + | No LOH | No LOH | LOH | NI | LOH |
| CP 243 | IDC grade 3 | 4.5 | 48 | + | LOH | No LOH | No LOH | NI | NI |
| CP 248 | IDC grade 3 | 4.0 | 47 | Unknown | NI | LOH | No LOH | No LOH | No LOH |
| CP 252 | IDC grade 3 | 2.8 | 90 | + | NI | No LOH | LOH | NI | LOH |
| CP 275 | IDC grade 3 | 5.2 | 45 | + | LOH | NI | NI | LOH | No LOH |
| CP 310 | IDC grade 3 | 1.7 | 53 | + | LOH | No LOH | No LOH | NI | LOH |
| CP 313 | IDC grade 3 | 5.5 | 49 | + | No LOH | NI | No LOH | NI | No LOH |
| CP 330 | IDC grade 3 | 2.0 | 43 | − | LOH | LOH | LOH | LOH | No LOH |
| CP 331 | IDC grade 3 | 4.2 | 55 | + | No LOH | No LOH | NI | No LOH | No LOH |
| CP 338 | IDC grade 3 | 3.5 | 42 | − | No LOH | NI | NI | No LOH | NI |
IDC, invasive ductal carcinoma; LOH, loss of heterozygosity; NI, not informative.
PCR amplification/LOH analysis
DNA from matched normal and tumour tissues was amplified by the polymerase chain reaction (PCR) using five microsatellite markers with dinucleotide repeats. The following markers were used: D3S1300, intragenic to FHIT on 3p14.2; D17S855, intragenic to BRCA1 on 17q21.2; D17S579 and D17S806 on 17q21.3; and D17S785 on 17q24. The markers were chosen based on their reported high heterozygosity rate.15 The forward primer for each set was labelled with either of two fluorescent dyes: HEX or FAM (Applied Biosystems/PE Biosystems, Foster City, California, USA).
In all cases, the markers were initially evaluated in the DNA from lymphocytes and the informative markers were studied in the tumour tissue. PCR was performed as described previously,16 using 20 ng/μl of genomic DNA, 100mM Tris/HCl, 500mM KCl, 480nM forward primer and 480nM reverse primer, 200μM dNTPs, and 0.5 unit of Taq polymerase in a total volume of 10 μl. Reactions were cycled as follows: 95°C for five minutes, then 35 cycles of 94°C for 15 seconds, 55°C for 15 seconds, and 72°C for 30 seconds, followed by final elongation at 72°C for 10 minutes.
Allele sizes were determined by electrophoresis of PCR products in 6% denaturing polyacrylamide gels and compared with ROX 500 size standards (Applied Biosystems), using an automated sequencer (ABI 377). The fluorescent signals from the alleles with different sizes were recorded and analysed using GENOTYPER version 2.1 and GENESCAN version 3.1 software (Applied Biosystems). After visual examination of computer printouts (by two independent observers), LOH was determined mathematically according to the Genotyper User Manual (Applied Biosystems).
Statistical analysis
Significance was determined using χ2 and t tests to evaluate whether LOH frequency at specific loci was equally distributed among the clinicopathological parameters; p values < 0.05 were considered significant.
RESULTS
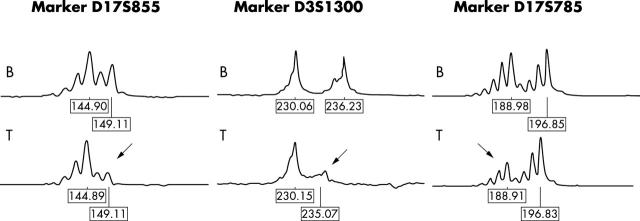
Specimens from 34 patients with sporadic invasive ductal carcinoma were assessed for LOH at the FHIT and the BRCA1 loci and at three other markers on 17q. Table 1 summarises the results. The BRCA1 intragenic marker, D17S855, showed the highest frequency of LOH (10 of 24 informative cases), followed by the D17S579 (six of 23 informative cases), D17S806 (five of 21 informative cases), and D17S785 markers (five of 21 informative cases). LOH at the FHIT intragenic marker, D3S1300, was found in six of 25 informative cases. Four of these six cases also had loss of the D17S855 marker. Figure 1 shows examples of losses at the BRCA1 and FHIT genes and at the D17S785 marker. There was no significant difference in the distribution of frequencies of samples that were non-informative, positive, or negative for LOH among the five microsatellite markers (χ28 = 4.64; p > 0.70).
Figure 1.
Examples of allelic losses (arrows) seen for the intragenic markers of the BRCA1 and FHIT genes and for the marker D17S785. B, blood; T, tumour.
DISCUSSION
Extensive searches for somatic mutations in the BRCA1 gene at 17q21 have revealed that such mutations are rare in non-familial breast and ovarian cancer, despite the high incidence of LOH at the BRCA1 locus seen in these tumours.6 Studies have suggested that loci surrounding the BRCA1 gene may be the major targets in 17q deletions.17
Frequent deletions in breast cancer are found on chromosome arm 3p, mainly at 3p14.2, where the FHIT gene is located,10–12 which can lead to LOH at this gene locus. High frequencies of LOH, abnormal transcripts, and reduced expression of the FHIT protein have been observed in invasive breast carcinomas, in addition to some benign breast diseases, suggesting that FHIT alterations are early events in mammary tumorigenesis.11,18,19
In our study, we have assessed LOH at 3p14 (FHIT gene) and three regions of chromosome 17q (17q21.2 (BRCA1 gene), 17q21.3, and 17q24) in patients with sporadic breast cancer. The highest incidence of LOH (42%) was found for the marker D17S855, intragenic to the BRCA1 gene. This finding is in agreement with other studies,5,15,20 supporting the hypothesis that the BRCA1 gene plays an important role in sporadic breast cancer. No significant difference was found for marker D17S855 between grade 2 and grade 3 tumours, suggesting that deletions in the BRCA1 gene probably occur early in mammary carcinogenesis. This result contrasts with data previously reported showing an association between LOH in BRCA1 and grade 3 tumours.21
Similar frequencies of LOH for markers D17S579 and D17S806 were found in other studies in sporadic breast cancer cases.15 The frequencies of LOH observed in this and other reports suggest the presence of other tumour suppressor genes at these regions that may participate in sporadic mammary tumorigenesis. However, we cannot rule out the possibility that the allelic losses in these regions may be a result of partial or total loss of chromosome 17, which is a common cytogenetic abnormality in breast cancer cases.
LOH for the marker D3S1300, intragenic to FHIT, has been reported to occur in frequencies ranging from 24% to 59% in breast cancer.10,13,22 In our study, we detected LOH at this marker in 24% of cases. Of interest was our observation that four of six cases that showed losses for the D3S1300 marker also showed losses at the BRCA1 marker, suggesting a possible association between allelic losses in these genes (table 1). It is possible that LOH at the BRCA1 gene occurs as an early event, causing genetic instability and leading to allelic losses at the FHIT region. Additional studies in larger series of cases using intragenic markers at these genes are necessary to test this hypothesis. However, a correlation between the expression of these genes was noted at the protein level by Turner et al.23 These authors found that cases with mutations of the BRCA1 gene also had reduced expression of the FHIT protein, suggesting the necessity of BRCA1 integrity to protect cells from genetic instability at fragile loci, such as FRA3B, where the FHIT gene is located.
“No significant difference was found for marker D17S855 between grade 2 and grade 3 tumours, suggesting that deletions in the BRCA1 gene probably occur early in mammary carcinogenesis”
The frequency of total LOH observed in the analysis of our sporadic breast tumour cases was found to be equally distributed among the tumours classified as grade 2 and grade 3. In addition, no significant difference was found when the LOH frequency was compared for tumour size (⩽ 2.8 cm and > 2.8 cm). However, when tumour size was compared between the two grades of tumours, it was found that grade 3 tumours were larger than grade 2 tumours.
Our study revealed that women ⩽ 50 years of age presented with tumours of larger size than women > 50 years, suggesting a faster tumour evolution in younger women. A higher frequency of LOH was also found in the younger group, although this observation did not reach significance by the χ2 test. Similar results were reported by Phelan and colleagues15 and Querzoli et al.24 Age and histopathological grades were not associated in this population, corroborating findings by Johnson et al.5 In addition, we found no increase of the total LOH frequency in grade 3 tumours compared with grade 2 tumours. Previous studies have reported that breast carcinoma in younger women (< 35 years old) usually presents with more aggressive pathological features, such as significantly higher proliferation rates, lack of steroid receptors, and a higher chance of having grade 3 tumours.25
In our cases, the tumours that presented with metastasis to the axillary lymph nodes did not have a higher frequency of LOH. This result is supported by other studies,15,22,26 but contrary to the study of Regitnig et al,25 who found an association between a higher frequency of LOH at the 17S855 marker and lymph node metastasis in a series of 40 primary sporadic breast tumours and their locally recurrent breast carcinomas.
Although it is important to evaluate breast cancer cases from different ethnic groups to look for the possible presence of unusual changes associated with specific ethnicities, we found no such differences at the markers studied here. Further studies using larger sporadic breast cancer samples and additional markers would be useful to confirm our findings and to establish more specific associations with clinicopathological parameters in our specific population of patients from Southern Brazil.
Take home messages.
The frequency of allelic losses in the FHIT and BRCA1 loci in the Southern Brazilian population is similar to that described in the general population
No correlations were found when the total loss of heterozygosity frequency was compared with tumour size, grade, or presence of axillary lymph node metastasis
Further studies using larger sporadic breast cancer samples and additional markers would be useful to confirm these findings, and to establish more specific associations with clinicopathological parameters in our patients
Acknowledgments
This work was partially supported by a research grant from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil). SSL was supported by a scholarship from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Ensino Superior, Brazil).
Abbreviations
LOH, loss of heterozygosity
PCR, polymerase chain reaction
REFERENCES
- 1.Ministério da Saúde. Instituto Nacional do Câncer (available at http://www.inca.gov.br). Last accessed on December 19, 2002.
- 2.Callahan R, Campbell G. Mutations in human breast cancer: an overview. J Natl Cancer Inst 1989;81:1780–6. [DOI] [PubMed] [Google Scholar]
- 3.Beckmann MW, Picard F, An HX, et al. Clinical impact of detection of loss of heterozygosity of BRCA1 and BRCA2 markers in sporadic breast cancer. Br J Cancer 1996;73:1220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanby AM, Kelsell DP, Potts HW, et al. Association between loss of heterozygosity of BRCA1 and BRCA2 and morphological attributes of sporadic breast cancer. Int J Cancer 2000;88:204–8. [PubMed] [Google Scholar]
- 5.Johnson SM, Shaw JA, Walker RA. Sporadic breast cancer in young women: prevalence of loss of heterozygosity at p53, BRCA1 and BRCA2. Int J Cancer 2002;98:205–9. [DOI] [PubMed] [Google Scholar]
- 6.Rosen EM, Fan S, Pestell RG, et al. BRCA1 gene in breast cancer. J Cell Physiol 2003;196:19–41. [DOI] [PubMed] [Google Scholar]
- 7.Van der Looij M, Cleton-Jansen AM, Van Eijk R, et al. A sporadic breast tumor with a somatically acquired complex genomic rearrangement in BRCA1. Genes Chromosomes Cancer 2000;27:295–302. [DOI] [PubMed] [Google Scholar]
- 8.Petersson C, Pandis N, Mertens F, et al. Chromosome aberrations in prophylactic mastectomies from women belonging to breast cancer families. Genes Chromosomes Cancer 1996;16:185–8. [DOI] [PubMed] [Google Scholar]
- 9.Glover TW, Stein CK. Chromosome breakage and recombination at fragile sites. Am J Hum Genet 1988;43:265–73. [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmadian M, Wistuba II, Fong KM, et al. Analysis of the FHIT gene and FRA3B region in sporadic breast cancer, preneoplastic lesions, and familial breast cancer probands. Cancer Res 1997;57:3664–8. [PubMed] [Google Scholar]
- 11.Campligio M, Pekarsky Y, Menard S, et al. FHIT loss of function in human primary breast cancer correlates with advanced stage of the disease. Cancer Res 1999;59:3866–9. [PubMed] [Google Scholar]
- 12.Gatalica Z, Lele SM, Rampy BA, et al. The expression of FHIT protein is related inversely to disease progression in patients with breast carcinomas. Cancer 2000;88:1378–83. [PubMed] [Google Scholar]
- 13.Ingvarsson S. FHIT alterations in breast cancer. Semin Cancer Biol 2001;11:361–6. [DOI] [PubMed] [Google Scholar]
- 14.Bergthorsson JT, Eiriksdottir G, Barkardottir RB, et al. Linkage analysis and allelic imbalance in human breast cancer kindreds using microsatellite markers from the short arm of chromosome 3. Hum Genet 1995;96:437–43. [DOI] [PubMed] [Google Scholar]
- 15.Phelan CM, Borg A, Cuny M, et al. Consortium study on 1280 breast carcinomas: allelic loss on chromosome 17 targets subregions associated with family history and clinical parameters. Cancer Res 1998;58:1004–12. [PubMed] [Google Scholar]
- 16.Cavalli LR, Singh B, Isaacs C, et al. Loss of heterozygosity in normal breast epithelial tissues and benign breast lesions in BRCA1/2 carriers with breast cancer. Cancer Genetics Cytogenetics 2004;149:38–43. [DOI] [PubMed] [Google Scholar]
- 17.Sourvinos G, Kiaris H, Tsikkinis A, et al. Microsatellite instability and loss of heterozygosity in primary breast tumours. Tumor Biol 1997;18:157–66. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi S, Tanimoto K, Hajiro-Nakanishi K, et al. Abnormal FHIT transcripts in human breast carcinomas: a clinicopathological and epidemiological analysis of 61 Japanese cases. Cancer Res 1997;57:1981–5. [PubMed] [Google Scholar]
- 19.Bieche I, Latil A, Becette V, et al. Study of FHIT transcripts in normal and malignant breast tissue. Genes Chromosomes Cancer 1998;23:292–9. [PubMed] [Google Scholar]
- 20.Ando Y, Iwase H, Ichihara S, et al. Loss of heterozygosity and microsatellite instability in ductal carcinoma in situ of the breast cancer. Cancer Lett 2000;156:207–14. [DOI] [PubMed] [Google Scholar]
- 21.Silva JM, Gonzalez R, Provencio M, et al. Loss of heterozygosity in BRCA1 and BRCA2 markers and high-grade malignancy in breast cancer. Breast Cancer Res Treat 1999;53:9–17. [DOI] [PubMed] [Google Scholar]
- 22.Maitra A, Wistuba II, Washington C, et al. High resolution chromosome 3p allelotyping of breast carcinomas and precursor lesions demonstrates frequent loss of heterozygosity and a discontinuous pattern of allele loss. Am J Pathol 2001;159:119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner BC, Ottey M, Zimonjic DB, et al. The fragile histidine triad/common chromosome fragile site 3B locus and repair-deficient cancers. Cancer Res 2002;62:4054–60. [PubMed] [Google Scholar]
- 24.Querzoli P, Albonico G, Di Lasio MG, et al. Biophenotypes and survival of BRCA1 and TP53 deleted breast cancer in young women. Breast Cancer Res Treat 2001;66:135–42. [DOI] [PubMed] [Google Scholar]
- 25.Regitnig P, Moser R, Thalhammer M, et al. Microsatellite analysis of breast carcinoma and corresponding local recurrences. J Pathol 2002;198:190–7. [DOI] [PubMed] [Google Scholar]
- 26.Takita K, Sato T, Miyagi M, et al. Correlation of loss of alleles on the short arms of chromosomes 11 and 17 with metastasis of primary breast cancer to lymph nodes. Cancer Res 1992;52:3914–17. [PubMed] [Google Scholar]