Abstract
Tim44 is an essential component of the machinery that mediates the translocation of nuclear-encoded proteins across the mitochondrial inner membrane. It functions as a membrane anchor for the ATP-driven protein import motor whose other subunits are the mitochondrial 70-kDa heat-shock protein (mhsp70) and its nucleotide exchange factor, mGrpE. To understand how this motor is anchored to the inner membrane, we have overexpressed Tim44 in Escherichia coli and studied the properties of the pure protein and its interaction with model lipid membranes. Limited proteolysis and analytical ultracentrifugation indicate that Tim44 is an elongated monomer with a stably folded C-terminal domain. The protein binds strongly to liposomes composed of phosphatidylcholine and cardiolipin but only weakly to liposomes containing phosphatidylcholine alone. Studies with phospholipid monolayers suggest that Tim44 binds to phospholipids of the mitochondrial inner membrane both by electrostatic interactions and by penetrating the polar head group region.
Import of nuclear-encoded proteins into the mitochondrial matrix is mediated by two protein complexes: the Tom complex in the mitochondrial outer membrane and the Tim complex in the mitochondrial inner membrane (1–3). Binding of a mitochondrial precursor protein to the Tom complex is stabilized by electrostatic interaction between the precursor’s positively charged presequence and the negatively charged domains of the Tom proteins (4–6). This charge interaction may also provide the initial driving force for the transport of precursor proteins across the mitochondrial outer membrane. The Tim complex mediates translocation across the inner mitochondrial membrane in a reaction requiring an electrochemical potential across the inner membrane (1–3). This potential provides the driving force for the insertion of the presequence into the Tim channel. Complete transport of the precursor into the matrix is then effected by an ATP-dependent reaction mediated by a transient heterooligomer composed of the three inner membrane proteins Tim23, Tim17, and Tim44, and the two matrix proteins mitochondrial hsp70 (mhsp70) and its nucleotide exchange factor, mitochondrial GrpE (mGrpE) (1–3; 7–10).
Genetic and biochemical studies on this ATP-dependent transport reaction have led to the following model (see refs. 1–3 for review): the import channel proper is formed by a 90-kDa heterooligomer consisting of Tim23 and Tim17 (11). During import of proteins into the matrix, the Tim17/23 channel may associate transiently with the Tom complex in the outer membrane and with Tim44 on the inner side of the inner membrane. Association with Tim44 recruits mhsp70 and mGrpE to the inner mouth of the Tim17/23 channel in a nucleotide-dependent manner. Thus, Tim44 functions as the membrane anchor for the ATP-driven import motor. The ATP-hydrolyzing subunit of this motor seems to be mhsp70, as it is the only motor subunit known to cleave ATP (7–10). Assembly and dissociation of the mhsp70–Tim44 complex are modulated by the nucleotide exchange factor mGrpE. Although the components of this motor have been identified, it is not yet clear how they work. In particular, it is not known how they are anchored to the inner membrane (12, 13) and how they interact with the Tim17/23 import channel. Data suggesting that Tim44 is a transmembrane protein imply that Tim44 is part of import channel, whereas data suggesting that Tim44 is a peripheral membrane protein imply a regulatory role in recruiting mhsp70 and mGrpE. In this report, we describe the purification and the structural analysis of Tim44 and the interaction of Tim44 with membrane phospholipids.
MATERIALS AND METHODS
N-Terminal Sequencing of Mature Tim44 from Yeast Mitochondria.
Purified yeast mitochondria (200 mg) containing hexahistidine-tagged Tim44 (8) were sonicated in 20 ml of buffer A [20 mM K-Hepes, pH 7.4/5% (vol/vol) glycerol/2 mM PMSF/1.25 μg/ml leupeptin/0.75 μg/ml antipain/0.25 μg/ml chymostatin/0.25 μg/ml elastin/5 μg/ml pepstatin] supplemented with 5 mM MgCl2 and 1 mM ATP and then centrifuged for 30 min at 20,000 × g. The pellet was solubilized in buffer A supplemented with 250 mM NaCl, 20 mM imidazole, 2 mM β-mercaptoethanol, and 0.6% of the nonionic detergent octylpolyoxyethylene. The solution was cleared by centrifugation, and the supernatant was incubated with Ni-nitrilotriacetic acid (NTA)-agarose for 2 h. Unbound proteins were washed away with buffer A, and Tim44 was eluted with 300 μl of 20 mM K-Hepes pH 7.4/100 mM NaCl/350 mM imidazole/0.5% octylpolyoxyethylene. The eluted material was applied to a reverse-phase HPLC column (2.1–30 mM Aquapore Butyl column) equilibrated with 0.1% trifluoroacetic acid. Tim44 was eluted with a linear gradient of 0.1% trifluoroacetic acid/60% (vol/vol) acetonitrile and subjected to N-terminal sequencing (14).
Construction of Hexahistidine-Tagged Tim44.
The TIM44 ORF was amplified by PCR with a forward primer (base pairs 126–150) and a reverse primer (base pairs 1,266–1,293) of the TIM44 ORF (15). The forward primer contained a restriction site for AflIII upstream of the coding sequence, and the reverse primer contained a restriction site for BamHI downstream of the coding sequence. A hexahistidine tag was added to the C terminus of Tim44 by using plasmid pQE60 (Qiagen, Chatsworth, CA). The PCR product was cut with AflIII and BamHI, and the plasmid was cut with NcoI (compatible with AflIII) and BamHI. The purified products were ligated with T4 DNA ligase (Boehringer Mannheim). The resulting recombinant plasmid pTim44–6His encodes a Tim44 protein in which the mitochondrial targeting sequence is replaced by Met-Ser and in which the C terminus is extended by Gly-Ser-Arg-Ser-His6. The construct was sequenced to confirm proper in-frame ligation and the fidelity of the Taq polymerase. The his-tagged Tim44 was overexpressed in Escherichia coli strain M15.
Purification of Hexahistidine-Tagged Tim44.
Bacterial transformants were grown and induced essentially as described (16). The cells (1 liter of liquid culture) were harvested, suspended in 40 ml of buffer B [50 mM Tris⋅HCl, pH 7.4/1% Triton X-100/3.2 M urea/300 mM NaCl/10 mM imidazole/10% (vol/vol) glycerol/2 mM β-mercaptoethanol and protease inhibitors as in buffer A], disrupted by sonication, and then centrifuged at 20,000 × g to clear the solution. The supernatant was incubated with 2 ml of Ni-NTA-agarose for 90 min at 4°C. Unbound proteins were removed by washing the beads three times with 15 ml of buffer B containing 2 mM PMSF. Bound Tim44 was then eluted with 7 ml of 50 mM Tris⋅HCl, pH 7.4/350 mM imidazole/3.2 M urea/10% (vol/vol) glycerol/10 mM KCl.
To purify Tim44 further, the Ni-NTA-agarose eluate was diluted 20-fold with buffer C (20 mM Tris⋅HCl, pH 7.4/10 mM KCl/4 M urea/1 mM β-mercaptoethanol) and loaded onto a MonoQ column (HR5/5; Amersham Pharmacia) that had been preequilibrated with buffer C. The column was washed with 30 ml of buffer C, and bound proteins were eluted with 30 ml of a linear NaCl gradient (0–220 mM). Full-length Tim44 and its degradation product eluted at 90 mM and 130 mM NaCl, respectively. Fractions enriched in full-length Tim44 were pooled, diluted to 30 ml with buffer D [50 mM Tris⋅HCl, pH 7.4/1% Triton X-100/3.5 M urea/250 mM NaCl/10 mM imidazole/10% (wt/vol) glycerol/2 mM β-mercaptoethanol], bound to 1 ml of Ni-NTA-agarose, and incubated at 4°C for 1 h. Tim44 bound to the Ni-NTA-agarose was washed sequentially with 15-ml aliquots of buffer D containing decreasing concentrations of urea and Triton (a decrease of 0.5 M urea and 0.25% Triton X-100 for each wash). Elution from the beads was achieved with 4 ml of 20 mM Tris⋅HCl, pH 7.4/350 mM imidazole/10% (wt/vol) glycerol. The eluted pure Tim44 was dialyzed against 20 mM Tris⋅HCl, pH 7.4/10% (wt/vol) glycerol/200 mM NaCl, concentrated to about 1 mg/ml in Centricon tubes (Amicon), frozen in liquid nitrogen, and stored at −80°C. Unless otherwise indicated, all studies on Tim44 described in this manuscript were carried out in the absence of detergents.
Proteolysis of Tim44.
Tim44 (0.2 mg/ml) was incubated for 2 min at 4°C with either trypsin or proteinase K in 20 mM K-Hepes, pH 7.4/100 mM KCl/10 mM MgCl2. Proteolysis was stopped by adding PMSF (2 mM) and electrophoresis sample buffer to 1% SDS/1% β-mercaptoethanol and by boiling for 5 min. Reaction mixtures (50 μl) were analyzed by SDS/14% PAGE.
Tim44 Binding to Sucrose-Loaded Vesicles.
Sucrose-loaded vesicles composed of either a mixture of cardiolipin (CL; from beef heart mitochondria) and dioleoyl phosphatidylcholine (PC; 1:5 wt/wt) or PC alone were prepared by using published protocols (17). Tim44 (0.044 mg/ml) was incubated at 22°C for 1 h in 200 μl of 20 mM K-Hepes, pH 7.4/100 mM NaCl. Tim44 bound to the vesicles was separated from unbound Tim44 by centrifugation at 55,000 rpm in a Beckman Optima TLX table-top ultracentrifuge (TLA-100.3 rotor) for 60 min at 22°C. The supernatant and pellet fractions were analyzed by SDS/12% PAGE (18). Bound Tim44 was determined by optical densitometry of Coomassie blue-stained bands obtained after SDS/PAGE analysis of the vesicle pellet (18).
Trypsinolysis of Vesicle-Bound Tim44.
Tim44 (0.044 mg/ml) was incubated at 20°C in the presence or absence of either CL/PC or PC vesicles in a 150-μl reaction mixture for 1 h. Next, trypsin was added, and the mixture was incubated for 2 min at 20°C. Proteolysis was stopped by the addition of PMSF to 2 mM and of electrophoresis sample buffer to 1% SDS/1% β-mercaptoethanol and by boiling for 5 min. The products of trypsinolysis were analyzed by SDS/14% PAGE.
Analytical Ultracentrifugation.
Sedimentation velocity and sedimentation equilibrium measurements were performed on Tim44 (0.4 mg/ml) at 20°C in 20 mM Na-Hepes with 150 mM NaCl as described (14).
Monolayer Experiments.
Tim44 penetration into the monolayer was followed by measuring the surface-pressure increase of the monolayer as described (19). The experiments were performed at room temperature and, unless stated otherwise, in 20 mM Tris⋅HCl (pH 7.4) containing 100 mM NaCl.
RESULTS
The N Terminus of Mature Yeast Tim44.
Tim44 is made as a precursor that is cleaved on import into mitochondria. To express mature Tim44 in bacteria, it was necessary to determine the N terminus of the mature protein. For this purpose, we purified mitochondria from a yeast strain that expressed Tim44 with a C-terminal hexahistidine tag. The tagged Tim44 protein was purified in two steps, first on Ni-agarose beads and then by reverse-phase HPLC (14). The N-terminal sequence of pure Tim44 was H2N-Glu-Gly-Gly-Asn-Pro-Arg-Ser-Pro. Comparison of this sequence with the nucleotide sequence of the TIM44 gene indicated that the presequence is cleaved after amino acid 42. However, although our analysis yielded Glu as the N-terminal residue of mature Tim44, the gene sequence predicted Gln. This discrepancy may be an artifact of the N-terminal sequence analysis or the result of a side reaction of cleavage by the matrix-processing peptidase, as we encountered the same discrepancy when sequencing yeast cytochrome oxidase subunit iv (20).
Properties of Purified Recombinant Tim44.
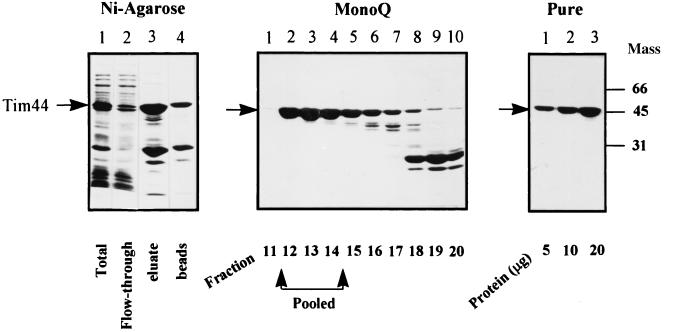
Previous complementation studies showed that the hexahistidine-tagged Tim44 complements a Tim44 disruption and thus functions similarly to wild-type Tim44 in vivo (9). To obtain large amounts of the protein, we therefore overexpressed the mature tagged Tim44 in Escherichia coli. Our initial attempts to purify this overexpressed Tim44 were unsuccessful, as the protein was extensively degraded. Degradation could be partially overcome by performing the two purification steps in the presence of urea. In this way, Tim44 was recovered from the Ni-NTA-agarose beads as two species, which were identified by immunoblotting and N-terminal sequencing as full-length Tim44 and its degradation product (Fig. 1A). These two species were successfully separated from each other by anion-exchange chromatography (Fig. 1B). Finally, urea was removed by rebinding the Tim44 to Ni-agarose and washing the beads sequentially with buffers containing decreasing concentrations of urea. Tim44 that eluted from the beads was >95% pure, as determined by SDS/PAGE (Fig. 1C). The purified Tim44 was equally soluble in buffers with or without nonionic detergents (not shown).
Figure 1.
Purification of recombinant yeast Tim44. (Left) Purification on Ni-NTA-agarose. Lane 1, a lysate of bacteria overexpressing Tim44; lane 2, proteins not bound to Ni-NTA-agarose; lane 3, proteins eluted from the Ni-NTA-agarose with imidazole; lane 4, proteins remaining on the beads after imidazole elution. (Center) Anion-exchange chromatography on a MonoQ column. Fractions enriched in Tim44 (5 μl) were analyzed by SDS/PAGE. (Right) Pure Tim44 was loaded in lanes 1–3 (5, 10, or 20 μg, respectively). Proteins were stained with Coomassie blue. Mass, molecular mass.
Circular-dichroism analysis of the purified Tim44 (not shown) identified mainly α-helices, suggesting that the purified recombinant protein was properly folded. When purified Tim44 was analyzed in the analytical ultracentrifuge by sedimentation to equilibrium, the calculated mass was 44 kDa. The purified protein is thus a monomer. Sedimentation velocity centrifugation yielded a sedimentation coefficient of 2.4 S. This low value indicates that the protein has an elongated shape (14).
Domain Organization of Tim44.
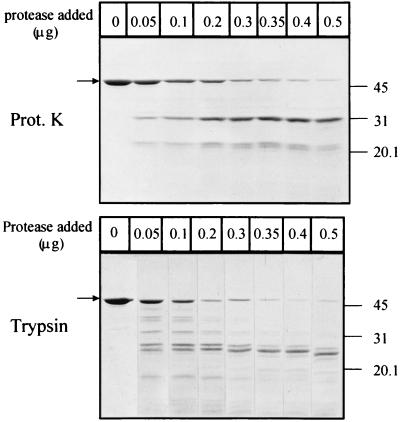
To define the domain organization of Tim44, we subjected the protein to limited proteolysis with proteinase K and trypsin (Fig. 2). The rationale behind this classical method is that proteases preferentially cleave within solvent-exposed, unstructured regions rather than within folded domains (21). Proteinase K generated essentially a single major fragment whose mobility on SDS/PAGE was consistent with an apparent mass of 31 kDa. N-terminal sequencing and mass spectroscopy suggested that this stable Tim44 domain starts at amino acid 210 and extends until the end of the C terminus of the protein. The predicted mass of this fragment is thus 26,685 kDa. Digestion of Tim44 with low trypsin levels yielded several products (Fig. 2); more extensive digestion gave a single major fragment. This fragment migrated with an apparent mass of 24.5 kDa on SDS/PAGE and still bound to Ni-NTA-agarose beads (not shown), indicating that it represented the C-terminal part of Tim44. We conclude that Tim44 contains a tightly folded 25-kDa domain at its C terminus.
Figure 2.
Proteolysis of pure Tim44. Tim44 was digested with either proteinase K or trypsin for 2 min, and the products of proteolysis were separated by SDS/14% PAGE. The amount of proteinase K and trypsin added to each 50-μl reaction mixture is indicated at the top. The arrow indicates the position of full-length Tim44. The molecular mass (in kDa) of the standards is indicated on the right.
Interaction of Tim44 with Model Membranes.
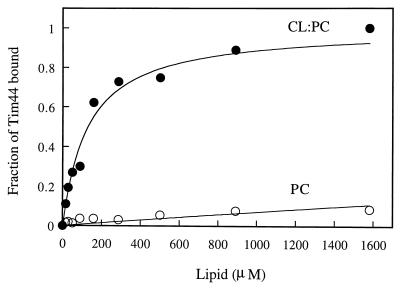
Interaction of Tim44 with phospholipid vesicles. To study the interaction of Tim44 with membranes, we used as a model system sucrose-filled phospholipid vesicles composed of either PC or a mixture of PC and CL. CL is the major acidic phospholipid of the mitochondrial inner membrane, with a net charge of −2 at physiological pH. The sucrose-filled vesicles were incubated with pure Tim44, collected by centrifugation, and analyzed for bound Tim44 by SDS/PAGE. Fig. 3 shows that Tim44 associated with the CL/PC vesicles as a function of the lipid concentration; half-maximal binding occurred at 129 μM lipid. This high affinity suggests that the observed association of Tim44 with CL/PC vesicles is specific. Maximal binding to PC vesicles was 10-fold lower than to PC/CL vesicles, and half-maximal binding occurred only at the 100-fold higher lipid concentration of 13 mM. These results suggest that Tim44 is not an integral membrane protein but that it binds to membranes by an interaction with acidic phospholipids.
Figure 3.
Binding of Tim44 to phospholipid vesicles. Pure Tim44 was incubated with vesicles composed of either CL and PC (1:5, wt/wt) or PC alone. The vesicles were collected by centrifugation, and protein associated with vesicles was detected by SDS/14% PAGE and staining with Coomassie blue. Bound Tim44 was quantified by densitometric integration of the peaks. Background “binding” (≈5% of Tim44 added without lipids) was subtracted, and the highest amount of vesicle-bound Tim44 (≈90% of Tim44 added) was taken as 100%. The continuous line represents the best fit of the data, assuming a dissociation constant of 129 μM and 13 mM for binding to PC/CL vesicles ● or PC vesicles ○, respectively.
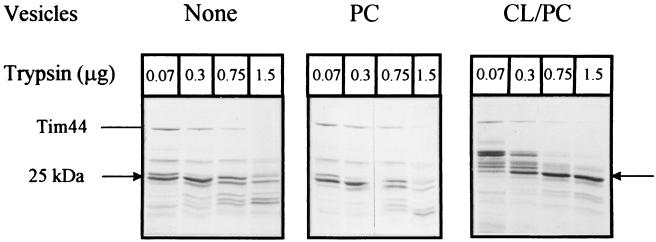
To test whether part of Tim44 inserts into the phospholipid bilayer and thereby becomes inaccessible to added protease, we bound Tim44 to PC or CL/PC vesicles, added trypsin to the suspending medium, and analyzed the resulting Tim44 fragments by SDS/PAGE (Fig. 4). With PC vesicles, we observed essentially the same digestion pattern as in the absence of vesicles. Under the conditions of this experiment, the stable domain was degraded by more than 0.3 μg of trypsin. In the presence of CL/PC vesicles, however, this domain was resistant to 1.5 μg of trypsin and retained its ability to bind CL/PC liposomes as well as Ni-NTA-agarose beads (not shown). These results indicate that Tim44 interacts with acidic phospholipids through its stable C-terminal domain.
Figure 4.
Trypsinolysis of vesicle-bound Tim44. Pure Tim44 was incubated with no vesicles (Left), PC vesicles (Center), or CL/PC vesicles (Right). The indicated concentrations of trypsin were added, and the proteolysis products were analyzed by SDS/PAGE and staining with Coomassie blue.
Interaction of Tim44 with phospholipid monolayers.
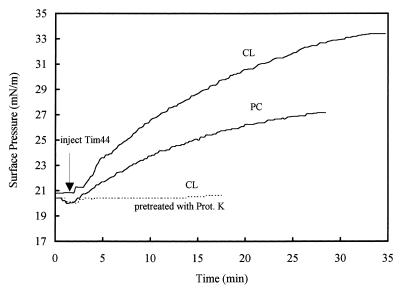
The experiments with vesicles were complemented by studying Tim44 binding to a phospholipid monolayer. Injection of Tim44 into the aqueous subphase caused the surface pressure of the monolayer to increase, suggesting that Tim44 penetrates into the monolayer (Fig. 5). At an initial surface pressure of 20 mN/m, the increase in surface pressure observed with a CL monolayer was twice that observed with a PC monolayer. No increase in surface pressure was observed when Tim44 was pretreated with proteinase K, indicating that the observed surface-pressure rise reflected association of intact Tim44 with the monolayer.
Figure 5.
Interaction of Tim44 with CL or PC monolayers. The initial surface pressure of both monolayers was adjusted to ≈20 mN/m. Pure Tim44 (0.8 mg/ml, 10 μl) was injected into the subphase, and the change in surface pressure was monitored as a function of time. (Dashed line) Tim44 was pretreated for 20 min with 6 μg of proteinase K before being injected into the subphase.
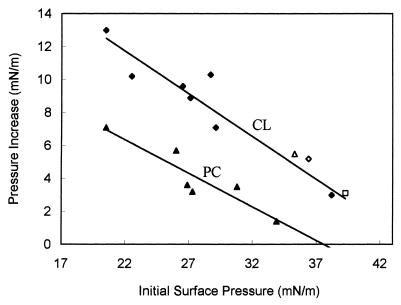
When we measured the Tim44-induced increase in surface pressure as a function of the initial surface pressure, we observed an inverse linear relationship (Fig. 6). However, at all initial surface pressures tested, the Tim44-induced surface-pressure increase was higher for CL monolayers than for PC monolayers. Extrapolation to zero pressure increase yielded significantly different limiting surface pressures for the two types of monolayers: association of Tim44 with a PC monolayer became undetectable at an initial pressure of 37.6 mN/m, whereas the corresponding value for a CL monolayers was 44.7 mN/m. As the lipid-packing density of the liposomes used in this study was equivalent to a monolayer surface pressure of 32–35 mN/m (22), the minimal binding of Tim44 observed to PC liposomes is readily explained. Strikingly, NaCl concentrations of up to 1 M did not affect the association of Tim44 with CL monolayers (Fig. 6).
Figure 6.
Tim44-induced surface-pressure increase as a function of initial surface pressure. The initial surface pressure of monolayers composed of CL (♦, ⋄, ▵, and □) or PC (▴) was set to the indicated values, and the change in surface pressure after Tim44 injection was recorded. Surface-pressure increase = (final surface pressure) − (initial surface pressure). Experiments were carried out at 100 mM (♦, ▴), 350 mM (⋄), 500 mM (▵), and 1 M NaCl (□).
DISCUSSION
The aim of this study was to characterize the association of Tim44 with membranes. We present several results that suggest that Tim44 is a peripheral membrane protein. First, Tim44 associates much more avidly with CL-containing model membranes than with model membranes containing only PC. Second, a part of the Tim44 molecule is protected from digestion when Tim44 is incubated with CL-containing liposomes; no such protection is seen with liposomes composed of only PC. Third, Tim44 is equally soluble in the presence and absence of nonionic detergents. Preferential binding of Tim44 to CL-containing vesicles and monolayers suggests that this binding is mediated by a charge interaction. However, Tim44 significantly increases the surface pressure of neutral PC monolayers, and addition of 1 M NaCl to the binding buffer does not prevent binding of Tim44 to CL monolayers. Both of the observations indicate that Tim44 also penetrates the polar head group region and interacts with the hydrophobic portion of the phospholipids.
We previously suggested that Tim44 spans the mitochondrial inner membrane (9). This conclusion was based on our observation that, in mitoplasts, Tim44 is partially accessible to antibodies in the suspending medium. Although the present results cast doubt on this earlier conclusion, it is also possible that association of Tim44 with the inner membrane is dynamic. We are currently testing the possibility that part of the protein is transiently exposed on the trans-side of the membrane, as has been reported for bacterial SecA (23).
Our data strongly argue against the possibility that Tim44 is part of the import channel proper. The major role of Tim44 may be to recruit mhsp70 and, indirectly, mGrpE to the inner mouth of the import channel. Modulation of membrane protein activity by its association with CL has been well documented (24). Therefore, it is tempting to speculate that the association of Tim44 with CL may serve as an initial step to anchor the ATP-driven import motor close to the import channel.
Acknowledgments
We thank Ariel Lustig for his expert help in carrying out the analytical ultracentrifugation studies. This work was supported by grants from the Swiss National Science Foundation (to G.S.), the Human Capital and Mobility Program of the European Economic Community (to G.S. and B.deK.), the Louis-Jeantet Foundation (to G.S.), and the Human Frontier Program Organization (to G.S.), as well as a Maof start-up fellowship (to A.A.) and a postdoctoral fellowship from Tel-Aviv University (to C.W.).
ABBREVIATIONS
- NTA
nitrilotriacetic acid
- PC
phosphatidylcholine
- CL
cardiolipin
References
- 1.Schatz G, Dobberstein B. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 2.Neupert W. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 3.Pfanner N, Meijer M. Curr Biol. 1997;7:100–103. doi: 10.1016/s0960-9822(06)00048-0. [DOI] [PubMed] [Google Scholar]
- 4.Dietmeier K, Hönlinger A, Bömer U, Dekker P J, Eckerskorn C, Lottspeich F, Kübrich M, Pfanner N. Nature (London) 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- 5.Komiya T, Rospert S, Koehler C, Looser R, Schatz G, Mihara K. EMBO J. 1998;17:3886–3898. doi: 10.1093/emboj/17.14.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schatz G. Nature (London) 1997;388:121–122. doi: 10.1038/40510. [DOI] [PubMed] [Google Scholar]
- 7.Schneider H-C, Berthold J, Bauer M F, Dietmeier K, Guiard B, Brunner M, Neupert W. Nature (London) 1994;371:768–774. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- 8.Berthold J, Bauer M F, Schneider H-C, Klaus C, Dietmeier K, Neupert W, Brunner M. Cell. 1995;81:1085–1093. doi: 10.1016/s0092-8674(05)80013-3. [DOI] [PubMed] [Google Scholar]
- 9.Kronidou N G, Oppliger W, Bolliger L, Hannavy K, Glick B S, Schatz G, Horst M. Proc Natl Acad Sci USA. 1994;91:12818–12822. doi: 10.1073/pnas.91.26.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rassow J, Maarse A C, Krainer E, Kübrich M, Müller H, Meijer M, Craig E A, Pfanner N. J Cell Biol. 1994;127:1547–1556. doi: 10.1083/jcb.127.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dekker P J, Martin F, Maarse A C, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N. EMBO J. 1997;16:5408–5419. doi: 10.1093/emboj/16.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horst M, Jenö P, Kronidou N G, Bolliger L, Oppliger W, Scherer P, Manning-Krieg U, Jascur T, Schatz G. EMBO J. 1993;12:3035–3041. doi: 10.1002/j.1460-2075.1993.tb05972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blom J, Kübrich M, Rassow J, Voos W, Dekker P J, Maarse A C, Meijer M, Pfanner N. Mol Cell Biol. 1993;13:7364–7371. doi: 10.1128/mcb.13.12.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azem A, Oppliger W, Lustig A, Jenö P, Feifel B, Schatz G, Horst M. J Biol Chem. 1997;272:20901–20906. doi: 10.1074/jbc.272.33.20901. [DOI] [PubMed] [Google Scholar]
- 15.Maarse A C, Blom J, Grivell L A, Meijer M. EMBO J. 1992;11:3619–3628. doi: 10.1002/j.1460-2075.1992.tb05446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horst M, Oppliger W, Rospert S, Schönfeld H-J, Schatz G, Azem A. EMBO J. 1997;16:1842–1849. doi: 10.1093/emboj/16.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buser C A, Sigal C T, Resh M D, McLaughlin S. Biochemistry. 1994;33:13093–13101. doi: 10.1021/bi00248a019. [DOI] [PubMed] [Google Scholar]
- 18.Vergères G, Manenti S, Weber T, Stürzinger C. J Biol Chem. 1995;270:19879–19887. doi: 10.1074/jbc.270.34.19879. [DOI] [PubMed] [Google Scholar]
- 19.Pilon M, Jordi W, de Kruijff B, Demel R A. Biochim Biophys Acta. 1987;902:207–216. doi: 10.1016/0005-2736(87)90297-5. [DOI] [PubMed] [Google Scholar]
- 20.Maarse A C, van Loon A P G M, Riezman H, Gregor I, Schatz G, Grivell L A. EMBO J. 1984;3:2831–2837. doi: 10.1002/j.1460-2075.1984.tb02216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz T, Lowenhaupt K, Kim Y-G, Li L, Brown B A, II. J Biol Chem. 1999;274:2899–2906. doi: 10.1074/jbc.274.5.2899. [DOI] [PubMed] [Google Scholar]
- 22.Marsh D. Biochim Biophys Acta. 1996;1286:183–223. doi: 10.1016/s0304-4157(96)00009-3. [DOI] [PubMed] [Google Scholar]
- 23.Economou A. Mol Microbiol. 1998;27:511–518. doi: 10.1046/j.1365-2958.1998.00713.x. [DOI] [PubMed] [Google Scholar]
- 24.Hase M, Yoshimi T, Ishikawa Y, Ohba A, Guo L, Mima S, Makise M, Yamaguchi Y, Tsuchiya T, Mizushima T. J Biol Chem. 1998;273:28651–28656. doi: 10.1074/jbc.273.44.28651. [DOI] [PubMed] [Google Scholar]