Abstract
Background: Although rare in mainland Japan, classic Kaposi’s sarcoma (KS) is frequently reported in Okinawa, a subtropical island in southern Japan. Human herpesvirus 8 (HHV8) has been identified in the tumours and geographical differences occur.
Aim: To sequence HHV8 in classic and AIDS associated KS in Okinawa.
Materials/Methods: Eight classic KS cases, one AIDS associated KS, five granuloma pyogenicum cases, two inflammatory pseudotumours, two Castleman’s disease cases, one angiosarcoma, and one primary effusion lymphoma (PEL) were studied. As a control, HHV8 positive cultured PEL cells (TY-1) were used. The presence of HHV8 sequences was evaluated by PCR and in situ hybridisation. PCR products were sequenced.
Results: There were no histological differences among KS resulting from the different virus genotypes. HHV8 was detected in all cases of KS, in one PEL, and one granuloma pyogenicum. Eight classic KS cases and one granuloma pyogenicum were infected with HHV8 genotype II/C (K1 region) or subtype C (ORF26 region), which had a five amino acid deletion at K1 VR2 region. An AIDS associated KS and a PEL were infected with type I/A virus.
Conclusion: In Okinawa, classic KS cases and one granuloma pyogenicum case were infected with HHV8 genotype II/C, also classified as subtype C. AIDS associated KS and PEL were infected with a different HHV8 (genotype I/A), similar to that found in the USA. In Okinawa, HHV8 infection is more than four times higher than in mainland Japan, resulting in many cases of KS because of HHV8 genotype II/C infection.
Keywords: human herpesvirus 8 genotype, Kaposi’s sarcoma, granuloma pyogenicum, Castleman’s disease, primary effusion lymphoma
A new γ herpesvirus, named Kaposi’s sarcoma (KS) associated herpesvirus or human herpesvirus 8 (HHV8), was identified in KS tumours by Chang et al in 1994.1 HHV8 infection appears to be necessary for the development of KS.2–6 The routes of HHV8 transmission and acquisition are unknown, although epidemic and serological data suggest that sexual transmission may occur.7 Up to now, many molecular epidemiological studies have been reported and have revealed geographical differences in the virus genome.8 Zong et al classified the virus into three distinct subgroups (A–C) using polymorphism of open reading frame (ORF) 26.9 Furthermore, a highly variable ORF K1 gene has been analysed as a marker for HHV8 genotyping,10–13 and four major molecular genotypes have been reported. Recently, five major molecular genotypes of the K1 gene I–V,10 and A–E12 have been used worldwide. Hybrid use—for example, I/A—is also commonly accepted. Poole et al have reported that all these genotypes have the M or P subtype of ORF K15 alleles at the right hand side.14
“In mainland Japan, Kaposi’s sarcoma (KS) is extremely rare even in patients with AIDS, but in Okinawa, a subtropical island south of mainland Japan, considerable numbers of classic KS have been reported”
AIDS associated KS is the most common tumour in human immunodeficiency virus (HIV) infected individuals. Until the emergence of AIDS, KS was considered to be a rare disorder and only episodic occurrences in the Mediterranean were reported. This is referred to as classic KS. In mainland Japan, KS is extremely rare even in patients with AIDS, but in Okinawa, a subtropical island located between the East China Sea and Pacific Ocean, 2,000 km south of mainland Japan, considerable numbers of classic KS have been reported.15,16 Furthermore, in the patients with KS reported in mainland Japan, many Okinawan resident cases have been included.17 Previously, we have reported HHV8 infection in KS tumours in Okinawa, in addition to seroepidemiological studies in the general population in Okinawa and in Kumamoto, which is in the south of mainland Japan.18 In total, 11 (6.5%) of 170 Okinawa serum samples, which included a minimum of three from each sex in a five year period covering every age from 0 to 95 years were positive for anti-HHV8 antibody. The seropositive rate increased with age, but two children less than 5 years old who had received blood transfusions were positive. In contrast, in Kumamoto, in the south mainland, only 2% (one of 50) of the samples were positive. Separately, in mainland Japan, Katano and colleagues19 reported that the seroprevalence of HHV 8 was 1.4% in the general population, and Fujii and colleagues20 reported a prevalence of 0.2% in the HIV negative population. HHV8 has also been reported to be an important causative agent in primary effusion lymphoma (PEL),21,22 Castleman’s disease,23 and other tumours.24–28 Furthermore, a correlation between HHV8 subtypes and the aggressiveness of KS has been described.29 Here, we examine the prevalence of HHV8 in the above diseases in Okinawa in relation to the histology of the tumours and the genotype of the virus. This is the first report of HHV8 genotype analysis in KS and the detection of HHV8 in granuloma pyogenicum, inflammatory pseudotumour, Castleman’s disease, PEL, and angiosarcoma in Okinawa.
MATERIALS AND METHODS
Samples and cell lines
All samples were obtained from Okinawan residents. There were nine patients with KS (eight classic and one AIDS associated KS), five with granuloma pyogenicum (including one pregnant woman), two with Castleman’s disease, two with inflammatory pseudotumours of the lung, one with angiosarcoma, and one with PEL (RM-P1).22 Seven of the KS cases and some other samples were non-fixed fresh samples. All samples were also fixed in phosphate buffered 10% formalin. One further sample was from a cultured PEL cell line (RM-P1),22 established from an HIV free Okinawa patient, which was cultured in RPMI 1640 supplemented with 20% fetal bovine serum (MP Biomedicals, Morgan Irvine, California, USA) at 37°C in 5% CO2.
A cultured PEL cell line, TY-1,21 was used as an HHV8 positive control. TY-1, which was established from an HIV positive mainland Japanese patient, was provided by Dr Katano (Department of Pathology, National Institute of Infectious Diseases, Tokyo, Japan), and was cultured in RPMI 1640 supplemented with 20% fetal bovine serum at 37°C in 5% CO2.
Morphological investigation
Samples fixed in 10% phosphate buffered formalin were routinely processed in paraffin wax and sectioned at 4 μm. Haematoxylin and eosin staining and periodic acid Schiff staining were performed on these sections. For the immunohistochemical staining, dewaxed sections were pretreated with 3% H2O2 for 20 minutes, washed, and blocked with a non-immune goat serum for 30 minutes. Antibodies to vascular endothelial growth factor (VEGF) (diluted 1/100; Wako, Osaka, Japan), interleukin 6 (IL-6; diluted 1/100; Dako, Kyoto, Japan), CD34 (diluted 1/100; Dako), the oestrogen receptor (diluted 1/100; Dako), basic fibroblast growth factor (bFGF; diluted 1/100; Wako), and cyclin D1 (diluted 1/100; Dako) were used.
Slides were incubated with the primary antibodies for 20 minutes, washed three times, and incubated with biotinylated second antibody with avidin and biotinylated horseradish peroxidase complex (Dako). 3′, 3-Diaminobenzidine (Dako) was used as a chromogen.
For in situ hybridisation (ISH) analysis, 115 bp of HHV8 ORF72 was amplified by the polymerase chain reaction (PCR), according to the method of Reed and colleagues30 and Gómez-Román et al.24 The following primer set was used: 5′-CACCCTGAAACTCCAGGC-3′ and 5′-GATCCGATCCTCACATAGCG-3′. The PCR products were biotinylated (Bio Prime DNA labelling system; Invitrogen, Carlsbad, California, USA) according to the manufacturer’s instructions. The ISH procedures were the same as reported previously.24,30
Detection of HHV8 DNA from tumours and cultured cell lines
DNA was routinely extracted from the 19 tumour tissues and the two cultured PEL cell lines. The specimens were digested with proteinase K (Merck, Tokyo, Japan) (110 μg/ml) in 500mM Tris/HCl buffer, pH 7.5, containing 0.45% Tween 20 and 2.5mM MgCl2 at 37°C for 36 hours. The DNA was extracted twice using phenol/chloroform (the phenol was equilibrated with 1.0M Tris/HCl, pH 8.0, containing 0.1% quinolinol; the chloroform was a 24/1 (vol/vol) mixture of chloroform and isoamyl alcohol), then once more with chloroform. The DNA was precipitated with three times the volume of 100% ethanol at −20°C. All DNA samples were used immediately after extraction. The 110 bp β globin gene was detected (data not shown) in all DNA samples according to the method of Saiki et al,31 using their primers (PCO3 and PCO4).
PCR was used to detect the presence of HHV8 sequences. The HHV8 ORF 26, K1, gBN, gBC, and gHM genes were amplified according to the methods of Lasota and Miettinen27 and Meng et al,10 using their primers (tables 1 and 2). The P (prominent) and M (minor) allelic forms of the ORF K15 region at the right hand side of the genome were analysed using the primers sets: K14.1 (P), LGH2079 (5′-GAGATCACTCTCCAACCAC-3′) and LGH2033 (5′-GGAGTGCCTTCCGTATAG-3′); K14.1 (M)(450 bp), LGH2079 (5′-GAGATCACTCTCCAACCAC-3′) and LGH2506 (5′-CACAGTCACCTATGCTAG-3′); K15 (P) (580 bp), LGH2476 (5′-GCAGTGTTTTATTAACGTC-3′) and LGH2477 (5′-CAACCCCATTTACTTC-3′); K15 (M) (370 bp), LGH2473 (5′-CATGCAGCGAGCTTGAGA-3′) and LGH2474 (5′-CTTTGAGTACTGTTTGTC-3′), according to the method of Poole et al.14 To prevent false positive results, sample preparation and PCR were carried out by three authors independently (TK, KC, and TI). Furthermore, distilled water was used as a negative control, and no positive reaction was obtained.
Table 1.
Primers for PCR amplification of the HHV8 ORF 26 gene
| PCR round | Primer | Nucleotide position1 | |
| 1st | KS1 | 5′-AGCCGAAAGGATTCCACCATT-3′ | 987–1007 |
| KS2 | 5′-ATCCGTGTTGTCTACGTCCAG-3′ | 1200–1220 | |
| 2nd | KS3 | 5′-CTCGAATCCAACGGATTTGA-3′ | 1011–1030 |
| KS4 | 5′-AATGACACATTGGTGGTATA-3′ | 1161–1180 | |
| 1st | KS5 | 5′-TCCCTCTGACAACCTTCAGA-3′ | 895–915 |
| KS8 | 5′-AACAGCTGCTGCAGAATAGC-3′ | 1071–1090 | |
| 2nd | KS6 | 5′-AGCAACACCCAGCTAGCAGT-3′ | 951–970 |
| KS8 | 5′-AACAGCTGCTGCAGAATAGC-3′ | 1071–1090 | |
| 1st | KS5 | 5′-TCCCTCTGACAACCTTCAGA-3′ | 896–915 |
| KS7 | 5′-TCAAATCCGTTGGATTCGAG-3′ | 1011–1030 | |
| 2nd | KS6 | 5′-AGCAACACCCAGCTAGCAGT-3′ | 951–970 |
| KS7 | 5′-TCAAATCCGTTGGATTCGAG-3′ | 1011–1030 |
The primers were those used by Lasota and Miettinen.27
HHV8, human herpesvirus 8; ORF, open reading frame; PCR, polymerase chain reaction.
Table 2.
Primers for PCR amplification of HHV8 K1, gBN, gBC, and gHM genes
| Gene | PCR round | Primer | Nucleotide position32 |
| K1 | 1st | 5′-GGCCCTTGTGTAAACCTGT-3′ | 51–69 |
| 5′-AGTATCCGACCTCATAAAATG-3′ | 1081–1061 | ||
| 2nd | 5′-GACCTTGTTGGACATCCTGTA-3′ | 76–96 | |
| 5′-ACTGGTTGCGTATAGTCTTCC-3′ | 961–941 | ||
| gBN | 1st | 5′-GACCTGTACGCCCTTCTGTAC-3′ | 8653–8673 |
| 5′-ATAGAGTGGCACGGGTCTC-3′ | 9136–9118 | ||
| 2nd | 5′-GCCACCCTGGGGACTGTCAT-3′ | 8720–8739 | |
| 5′-TTGGTGATGGCGGACTCTGTC-3′ | 9102–9082 | ||
| gBC | 1st | 5′-CCTGGGTGGCATCGGAAAAAC-3′ | 10795–10815 |
| 5′-GCGTGGGTTGCCTCACAGTGT-3′ | 10432–11413 | ||
| 2nd | 5′-ATTGGTTACCGGATTCATAAA-3′ | 10858–10878 | |
| 5′-GGGTCGATAAATGGATTGA-3′ | 11391–11373 | ||
| gHM | 1st | 5′-GCGCTCTATGGAGTGGTGTC-3′ | 37689–37708 |
| 5′-CTAGAAAGCAGGGGGAGAATG-3′ | 38624–38605 | ||
| 2nd | 5′-GACGGCGTCCCATCTTCTGTT-3′ | 37760–37781 | |
| 5′-GGGCAGCTGTCGGTGAGG-3′ | 38473–38456 |
The primers used were those used by Meng et al.10
HHV8, human herpesvirus 8; PCR, polymerase chain reaction.
Sequencing of the PCR products
The PCR products were extracted from agarose gel and cloned into a T vector prepared from Bluescript (Stratagene, La Jolla, California, USA), according to the method of Marchuk et al.33 The Bluescript plasmid was digested with EcoRV, and incubated with Taq polymerase (Cetus-Takara, Otsu, Japan; 1 unit/μg plasmid/20 μl volume) using standard buffer conditions (10mM Tris/HCl, pH 8.3, 50mM KCl, 1.5mM MgCl2, and 200 μg/ml bovine serum albumin) in the presence of 2mM dTTP for two hours at 70°C. The sequence analysis was carried out using a Hitachi SQ 5500 DNA sequencer (Hitachi, Tokyo, Japan) and LI-COR DNA sequencer, Model 4200 (LI-COR, Lincoln, Nebraska, USA). We performed bidirectional sequencing of the PCR products at least four times for each clone.
RESULTS
Morphological examination
Table 3 lists the samples. The histology of the KS cases varied slightly according to the clinical stage, but fundamentally they showed the same features. Figure 1A shows a classic type KS lesion in a 57 year old man (case 5 in table 3). Figure 1B shows an AIDS associated plaque stage KS (case 9 in table 3) that showed a mixture of spindle cells and capillary spaces within the dermis. Cases 10–14 in table 3 were granuloma pyogenicum, and were well circumscribed, 0.5–1.0 cm in diameter, and exophytic, with a shallow ulcer at the centre. Numerous rounded anastomosing vessels with slightly plump endothelial cells were embedded within a markedly oedematous stroma (fig 2A; case 10 in table 3). Neutrophil and lymphocyte infiltration was prominent. However, in one case (fig 2B; case 12 in table 2), which occurred at the instep region of the left foot, detailed examination using serial sections revealed KS-like lesions in the limited peripheral areas. A lymphoangioma-like pattern (fig 2C) and vascular spaces dissecting collagen fibres (fig 2D) were seen. HHV8 was detected in this sample, whereas the other four cases of granuloma pyogenicum were negative for HHV8. No patients with granuloma pyogenicum had multiple lesions, and after resection there were no recurrent tumours. The two Castleman’s disease cases were of localised type showing a hyaline vascular pattern of histology (data not shown). The two cases of inflammatory pseudotumour of the lung (data not shown) were asymptomatic, and found by a routine radiological examination at the left S3 lobe and left S7 lobe, respectively. The tumours were composed of fibrous tissues with severe plasma cell and lymphocyte infiltration. They were considered to be plasma cell granuloma type inflammatory pseudotumours.
Table 3.
List of samples
| Case | Age | Sex | Histological diagnosis | HIV | HHV8 |
| 1 | 72 | M | Classic KS | − | + |
| 2 | 91 | F | Classic KS | − | + |
| 3 | 82 | M | Classic KS | − | + |
| 4 | 86 | M | Classic KS | − | + |
| 5 | 57 | M | Classic KS | − | + |
| 6 | 54 | M | Classic KS | − | + |
| 7 | 86 | M | Classic KS | − | + |
| 8 | 78 | F | Classic KS | − | + |
| 9 | 32 | M | AIDS KS | + | + |
| 10 | 61 | M | GP | − | − |
| 11 | 32 | F | GP, pregnancy | − | − |
| 12 | 81 | M | GP | − | + |
| 13 | 35 | F | GP | − | − |
| 14 | 33 | M | GP | − | − |
| 15 | 65 | F | IF | − | − |
| 16 | 59 | M | IF | − | − |
| 17 | 24 | M | CD | − | − |
| 18 | 49 | M | CD | − | − |
| 19 | 63 | M | PEL (RM-P1)* | − | + |
| 20 | 71 | M | AS | − | − |
| Control | 45 | M | AIDS PEL (TY-1) | + | + |
AS, angiosarcoma; CD, Castleman’s disease; GP, granuloma pyogenicum; HHV8, human herpesvirus 8; HIV, human immunodeficiency virus; IF, inflammatory pseudotumour; KS, Kaposi’s sarcoma; PEL, primary effusion lymphoma; +, infected; −, not infected.
*The lymphoma cells were positive for Epstein-Barr virus type A.
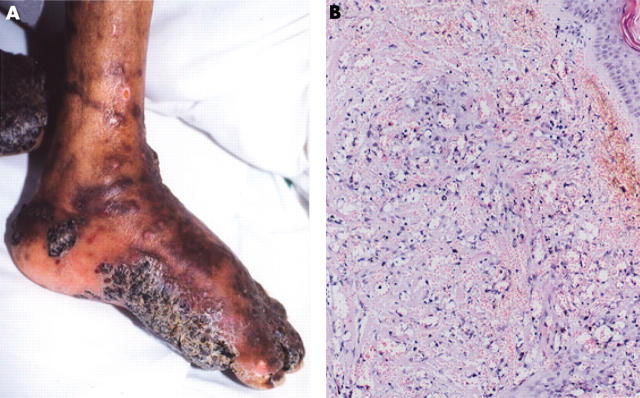
Figure 1.
(A) Advanced skin lesion in classic Kaposi’s sarcoma (KS) (case 5 in table 3). Many dark red/purple macules, plaques, and nodular lesions were seen and oedema was present. These lesions are often found in distal extremities. (B) Histology of AIDS associated KS (case 9 in table 3). A mixture of spindle cells and vascular spaces is seen. The vascular spaces are irregular and are dissected by bundles of collagen. Haematoxylin and eosin staining; original magnification, ×150.
Figure 2.
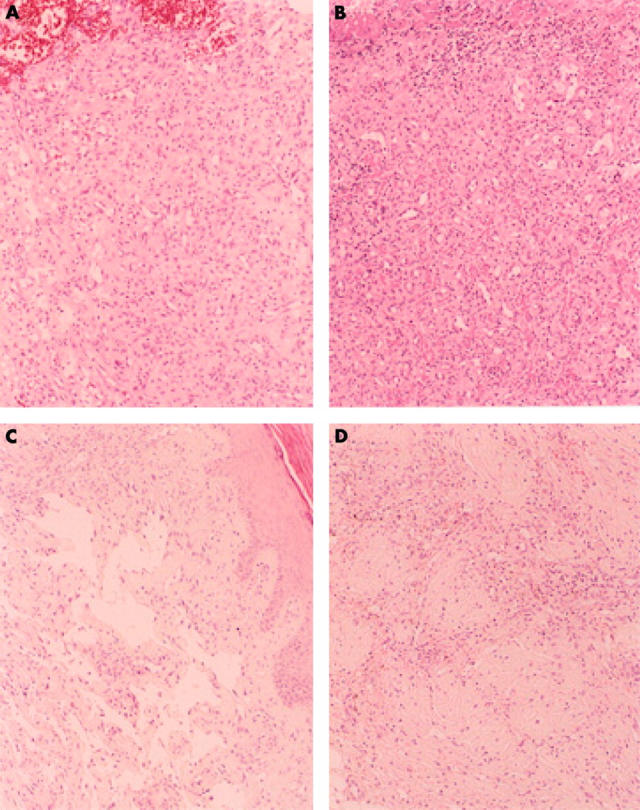
(A) Histology of granuloma pyogenicum (case 10 in table 3). Many rounded anastomosing small vessels with slightly plump endothelial cells were found within the oedematous stroma. Haematoxylin and eosin staining; original magnification, ×150. (B) Histological findings of granuloma pyogenicum (human herpesvirus 8 (HHV8) positive case; case 12 in table 3). Typical granuloma pyogenicum histology is noted, showing the anastomosing small vessel proliferation. Haematoxylin and eosin staining; original magnification, ×150. (C) Same case as in (B); in a small area, a lymphoangioma-like pattern is noted. Haematoxylin and eosin staining; original magnification, ×150. (D) Same case as in (B); in a small area, vascular spaces dissecting collagen bundles are seen. Haematoxylin and eosin staining; original magnification, ×150.
One of angiosarcomas was obtained from a 71 year old man. The tumour was a cutaneous angiosarcoma of the scalp and was composed of irregular vascular channels infiltrating the dermis.
Of the two PEL cases, TY-121 (the control case) was an AIDS associated case from a patient from mainland Japan, whereas RM-P122 was established from a non-AIDS Okinawan patient who had received haemodialysis for renal failure. The RM-P1 cells were positive for Epstein-Barr virus type A.
The spindle cells and vascular endothelial cells of KS and granuloma pyogenicum showed strong immunohistochemical staining for CD34, as did the tumour cells of the angiosarcoma. However, staining for VEGF, the oestrogen receptor, IL-6, and bFGF was negative in the granuloma pyogenicum cases, although VEGF, cyclin D1, and IL-6 were seen in a small number of the vascular endothelial cells and spindle cells in KS.
ISH revealed the presence of HHV8 mainly in the cytoplasm and a few nuclei of vascular endothelial cells and spindle cells of both classic and AIDS associated KS, as reported by Reed et al (fig 3).30 KS-like areas and some surrounding areas in one case of granuloma pyogenicum also showed a few positive signals. In KS, the ISH positive signals were more frequently seen in nodular lesions than in the patchy lesions. HHV8 was not detected in the granuloma pyogenicum lesions, although it was seen in PEL cells.
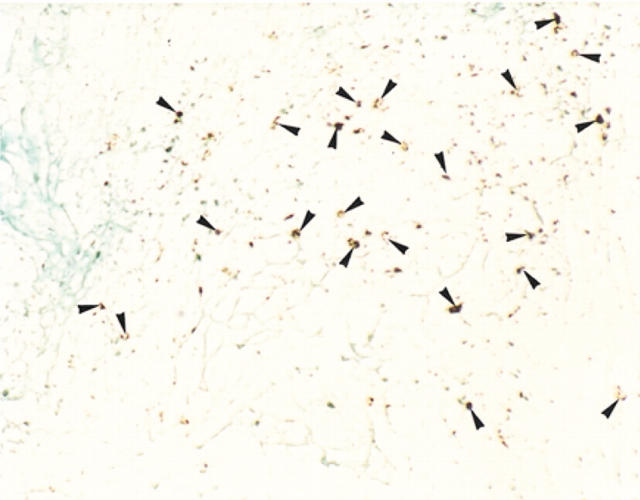
Figure 3.
Demonstration of human herpesvirus 8 (HHV8) in Kaposi’s sarcoma by in situ hybridisation. HHV8 signals (arrowheads) are found in the cytoplasm and a few nuclei of vascular endothelial cells and spindle cells (case 5 in table 3).
Detection of HHV8 and sequence analysis
HHV8 was detected in all KS samples. The two cases of PEL (RM-P1 and control TY-1) were also positive for HHV8. Among five cases of granuloma pyogenicum, one case (case 12 in table 3) was positive for HHV8. Table 4 summarises the results. All Okinawan resident KS cases were infected with HHV8 genotype II/C (analysis of K1 region),10–12 which is also classified as C subtype (analysis of ORF26 region),9 with the exception of one patient with AIDS associated KS, who had been infected with HIV in America. It is thought that this patient was infected with HHV8 at the same time. The HHV8 type detected in the two cases of PEL was also different from the Okinawan resident type. TY-1 was from a mainland Japanese patient who was also infected with HIV. RM-P1 was from an Okinawan patient, but the patient had received haemodialysis since the age of 50. He may have become infected with HHV8 through such treatment. The HHV8 positive granuloma pyogenicum case was classified as both genotype II/C (K1 region) and C (subtype of ORF26 region) virus. The virus detected in Okinawan residents was similarly classified into genotype II/C (K1 region) and subtype C (ORF26 region). There were no differences in the histology of the KS samples among the various HHV8 genotypes. The HHV8 infected granuloma pyogenicum case had KS-like lesions in a small area in addition to the typical granuloma pyogenicum histology.
Table 4.
Summary of the HHV8 genotype/subtype analysis
| Case | Tumour type | Genotype (K1 region) | Subtype (ORF26 region) | Subtype of M and P alleles |
| 1 | Classic KS | II/C | C | M |
| 2 | Classic KS | II/C | C | M |
| 3 | Classic KS | II/C | C | M |
| 4 | Classic KS | ND | C | M |
| 5 | Classic KS | ND | C | M |
| 6 | Classic KS | ND | C | M |
| 7 | Classic KS | ND | C | M |
| 8 | Classic KS | ND | C | M |
| 9 | AIDS KS | I/A | B | P |
| 10 | PEL | I/A | A | M |
| 11 | GP | II/C | C | M |
| Control | AIDS PEL | I/C | A | P |
Cases 1–11 are cases No. 1, 2, 3, 4, 5, 6, 7, 8, 9, 19 and 12 in table 3, respectively.
GP, granuloma pyogenicum; HHV8, human herpesvirus 8; KS, Kaposi’s sarcoma; ND, not determined; PEL, primary effusion lymphoma.
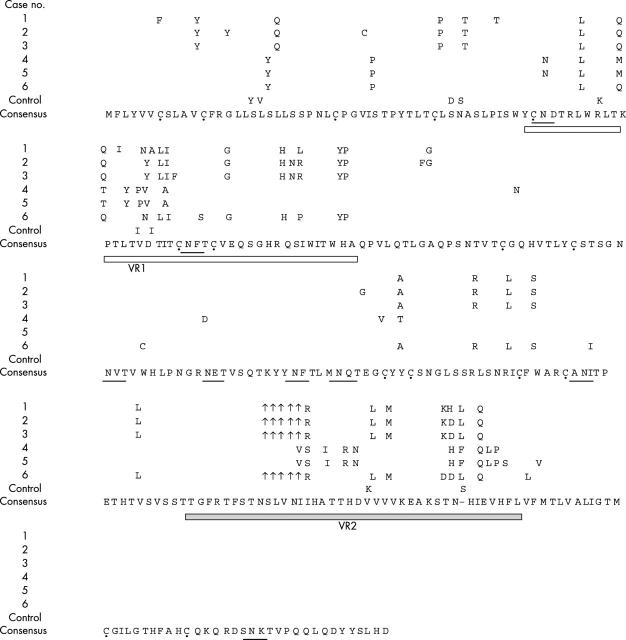
In the HHV8 K1 region, eight N-linked glycosylation sites were highly conserved in all cases, and cysteine residues were also well conserved, but in Okinawa HHV8, the amino acid at position 12 in three cases (cases 1, 2, and 3 in table 4 and fig 4) was tyrosine, that at position 7 in one case (case 1 in table 4 and fig 4) was phenylalanine, and at 71 in one other case (cases 3 in table 4 and fig 4) was also phenylalanine. In the variable region VR1 (amino acid positions 52–92) and VR2 (amino acid positions 191–231), there are many nucleotide variations and a five amino acid deletion is often seen in Okinawa genotype II/C HHV8, as shown in fig 4, although in other genotypes of HHV8 no such deletion was found.
Figure 4.
Alignment of K1 amino acid sequences of human herpesvirus 8 (HHV8). VR1 and VR2, variable regions 1 and 2. The dots below the sequence indicate cysteine residues. Underlining indicates potential N-linked glycosylation sites. Cases 1–3, classic Kaposi’s sarcoma (KS) (cases 1–3 in table 3); case 4, AIDS associated KS (case 9 in table 3); case 5, RM-P1 cells (case 19 in table 3); case 6, granuloma pyogenicum (case 12 in table 3); control, TY-1 cells.
In the C-terminal region, parts of the immunoreceptor tyrosine based activation motif (ITAM) was analysed, and was found to be conserved. Okinawa HHV8 also shows nucleotide variations at the ORF26 gene. The nucleotides at positions 981, 1032, 1055, 1086, 1132, and 1139 in Okinawa HHV8 ORF26 were C, C, G, T, A, and C, respectively, resulting in classification as subtype C according to Zong’s classification of the ORF26 region.9,12
The nucleotides at 981, 1032, 1055, 1086, and 1139 of the ORF26 gene of AIDS associated KS (case 9 in table 3) and both PELs (TY-1 and RM-P1) were C, A, T, C, G, and C and T, C, G, C, A, and A, respectively. They were thus classified as B and A subtypes, respectively.
The gBN and gHM segments were included in the gB and gH coding sequences. The gBN, gBC and gHM sequences were well conserved. At the nucleotide level, up to six variations were noted at position 383 of gBN (case 1 in table 3 showed a six nucleotide variation, but others showed none or only one nucleotide variation), one at position 534 of gBC, including 129 bp of non-translated region, and three to 10 at position 714 of gHM (case 1 in table 3 showed a 10 nucleotide variation, but others showed only three to four nucleotide variations). At the amino acid level, one variation was found at position 127 of gBN (case 1 in table 3), one at position 135 of gBC (cases 1 and 2 in table 3), and two to six at position 238 of gHM (cases 1 and 2 in table 3).
M and P allele analysis of Okinawan resident genotype II/C HHV8 and genotype I/A of the PEL (RM-P1) revealed the M type allele. The P type allele was detected in the genotype I viruses found in AIDS associated KS and control AIDS associated PEL.
Figure 4 and table 4 show the results of HHV8 sequence analysis.
DISCUSSION
In Okinawa, considerable numbers of KS cases have been reported, and all are infected with HHV8. As reported previously,18 the seroprevalence of HHV8 in Okinawa is 6.5% in the general population. Okinawa consists of many islands, but the high seropositivity is common among the islands. The HHV8 found in Okinawan residents was HHV8 type II/C (genotype of the K1 region) and C (subtype of the ORF26 region) according to the classifications of Meng and colleagues10,11 and Zong et al, respectively.9,12 However, in Okinawa type II/C HHV8, slight variations were found when compared with the reported sequences,1,10,32,34 although a five amino acid deletion at the VR2 region was also noted in a few HHV8 type II/C cases reported by Meng et al.10 In the Okinawan virus, amino acid sequence KDL is characteristically found in the VR2 and this is considered to resemble the KDEL sequence of the C-terminal of endoplasmic reticulum proteins. The ITAM sequence is thought to be conserved at the C-terminal region. Furthermore, the gB and gH sequences, which encode hypothetical immune target glycoproteins,10 are also well conserved. In contrast, the AIDS associated KS case in our study was infected with a different HHV8 type. In this patient, HIV infection had occurred in the USA, and infection with HHV8 is thought to have taken place at the same time. The HHV8 types in the two cases of PEL were also different from the Okinawan resident type. However, one of the granuloma pyogenicum cases was infected with Okinawan resident type HHV8. It is thought that in Okinawa, HHV8 genotype II/C/subtype C is prevalent in the general population.
Analysis of the HHV8 genotypes might clarify the route of virus infection. As has been reported previously, the two HHV8 genotypes I/A and II/C are worldwide predominant strains.11 In Asia, in Xinjiang in north western China, which is surrounded by Mongolia, Russia, Kazakhstan, Kyrgyz, and India, all KS cases are infected with genotype C (K1 region classification12) HHV8,35 and surprisingly, in Uygur people in Xinjiang, the seropositive rate of HHV8 is 46%.35 In mainland Japan, both strains (I/A and II/C) have been detected. Recently, Meng et al reported five Japanese KS cases where the HHV8 genotype was I/A and eight where it was II/C.11 It is thought that the patients with genotype I/A viruses had been infected in the USA, at the same time as being infected with HIV. Furthermore, in the Japanese in Hokkaido, northern Japan, another viral genotype has been noted. In the USA, according to the report of Meng et al,10 genotype I/A predominates over II/C.
Our present study has revealed that Okinawan resident classic KS cases are infected with genotype II/C virus, which is the M subtype of the ORF15 allelic form, but that the patient with AIDS associated KS, who was infected with HIV in the USA, harboured a genotype I/A and P subtype of the ORF15 allelic form of the virus. This led us to postulate that many classic KS cases in Okinawa are caused by genotype II/C HHV8 infection in the general population.
“The histology of the granuloma pyogenicum cases infected with HHV8 is slightly different to that of non-HHV 8 infected cases”
The histology of the KS cases does not differ fundamentally among the cases infected with different genotypes of the virus (II and I), although the possibility that strain differences are associated with different disease states has been suggested.12,26,29 The histology of the granuloma pyogenicum cases infected with HHV8 is slightly different to that of non-HHV 8 infected cases—HHV8 was demonstrated on the spindle cells and vascular endothelial cells in KS-like lesions, but not in the typical granuloma pyogenicum lesions. HHV8 was not detected in the inflammatory pseudotumours, Castleman’s disease cases, and the angiosarcoma in our series. However, the Castleman’s disease cases studied here were not of the multicentric type. In Okinawa, the seroprevalence of HHV8 is more than four times higher than that in mainland Japan,18–20 and KS is found more frequently in Okinawan residents than in mainland Japanese,15–17 but the numbers of Castleman’s disease and inflammatory pseudotumour cases in Okinawa are no higher than those in the mainland. The tumorigenic effect of HHV8 should be studied further. In addition, Katano et al reported that in KS lesions, latency associated nuclear antigen was found in large number of cells, and most cells showed latent infection.36 Therefore, study of the state of virus infection in KS is also needed.
Poole et al have reported that the K1 genes analysed previously can be categorised into four major subtypes with distinctive ethnic and geographical associations, which we believe have probably arisen during the migratory divergence of modern humans in Palaeolithic times.13 Therefore, it is also necessary to analyse HHV8 in the neighbouring countries in Asia and the islands of the South Pacific Ocean.
Take home messages.
Classic Kaposi’s sarcoma (KS) cases and one granuloma pyogenicum case in Okinawa were infected with human herpesvirus 8 (HHV8) genotype II/C, also classified as subtype C
AIDS associated KS and primary effusion lymphoma were infected with a different HHV8 (genotype I/A), similar to that found in the USA
In Okinawa, HHV8 infection is more than four times higher than in mainland Japan, resulting in a high prevalence of KS, although the numbers of inflammatory pseudotumours and Castleman’s disease cases are similar to that found in the mainland
There were no histological differences between the KS cases infected by the different genotypes
Dilnur et al have reported that Xinjiang, in China, is the most endemic area for HHV8 infection in the world known to date.35 Xinjiang is located at the middle point of the silk road and the virus may have been transmitted via the migration of people along this route. More than 100 years ago, Okinawa was an independent kingdom, and had close relations with China. The high incidence of KS and the high prevalence of HHV8 may reflect in part the previous history of the region.
Abbreviations
bFGF, basic fibroblast growth factor
HHV8, human herpesvirus 8
HIV, human immunodeficiency virus
IL-6, interleukin 6
ISH, in situ hybridisation
ITAM, immunoreceptor tyrosine based activation motif
KS, Kaposi’s sarcoma
ORF, open reading frame
PEL, primary effusion lymphoma
PCR, polymerase chain reaction
VEGF, vascular endothelial growth factor
VR, variable region
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994;266:1865–9. [DOI] [PubMed] [Google Scholar]
- 2.Cheng EH-Y, Nicholas J, Bellows DS, et al. A Bcl-2 homolog encoded by Kaposi sarcoma associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci U S A 1997;94:690–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li-M, Lee H, Yoon D - W, et al. Kaposi’s sarcoma-associated herpesvirus encodes a functional cyclin. J Virol 1997;71:1984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neipel F, Albrecht J-C, Ensser A, et al. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol 1997:839–42. [DOI] [PMC free article] [PubMed]
- 5.Neipel F, Albrechet J - C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol 1997;71:4148–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samaniego F, Pati S, Karp JE, et al. Human herpesvirus 8 K1-associated nuclear factor-kappa B-dependent promotor activity: role in Kaposi’s sarcoma inflammation? J Natl Cancer Inst Monogr 2000;28:15–23. [DOI] [PubMed] [Google Scholar]
- 7.Beral V, Peterman TA, Berkelman RL, et al. Kaposi’s sarcoma among persons with AIDS: a sexually transmitted infection? Lancet 1990;335:123–8. [DOI] [PubMed] [Google Scholar]
- 8.Huang L-M, Huang S-Y, Chen MY, et al. Geographical differences in human herpesvirus 8 seroepidemiology: a surgery of 1, 201 individuals in Asia. J Med Virol 2000;60:290–3. [PubMed] [Google Scholar]
- 9.Zong J-C, Metroka C, Reitz MS, et al. Strain variability among Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genomes: evidence that a large cohort of United States AIDS patients may have been infected by a single common isolate. J Virol 1997;71:2505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng Y-X, Spira TJ, Bhat GJ, et al. Individuals from North America, Australasia, and Africa are infected with four different genotypes of human herpesvirus 8. Virology 1999;261:106–119. [DOI] [PubMed] [Google Scholar]
- 11.Meng X-Y, Sata T, Stamey FR, et al. Molecular characterization of strains of human herpesvirus 8 from Japan, Argentina and Kuwait. J Gen Virol 2001;82:499–506. [DOI] [PubMed] [Google Scholar]
- 12.Zong J-C, Ciufo DM, Alcendor DL, et al. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi’s sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol 1999;73:4156–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bigger RJ, Whiteby D, Masshall V, et al. Human herpesvirus 8 in Brazilian Amerindians: a hyperendemic population with a new subtype. J Infect Dis 2000;181:1562–8. [DOI] [PubMed] [Google Scholar]
- 14.Poole LJ, Zong J-C, Ciufo DM, et al. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi’s sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J Virol 1999;73:6646–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwamasa T, Chinen K, Hirayasu T, et al. Epidemic and non-epidemic Kaposi’s sarcoma: diagnosis, staging and treatment. Crit Rev Oncol Hematol 1996;24:153–63. [DOI] [PubMed] [Google Scholar]
- 16.Kamada Y, Iwamasa T, Miyazato M, et al. Kaposi’s sarcoma in Okinawa. Cancer 1992;70:861–8. [DOI] [PubMed] [Google Scholar]
- 17.Hisaoka M, Hashimoto H, Iwamasa T. Diagnostic implication of Kaposi’s sarcoma-associated herpesvirus with special reference to the distinction between spindle cell hemangioendothelioma and Kaposi’s sarcoma. Arch Pathol Lab Med 1998;122:72–76. [PubMed] [Google Scholar]
- 18.Higa M, Iwamasa T, Sunakawa H. Seroprevalence of human herpesvirus 8 (HHV 8) in general population in Okinawa, a subtropical island in Japan. Proceeding of the Japanese Society of Pathology 2001;90:348. [Google Scholar]
- 19.Katano H, Iwasaki T, Baba N, et al. Identification of antigenic proteins encoded by human herpesvirus and seroprevalence in the general population and among patients with and without Kaposi’s sarcoma. J Virol 2000;74:3478–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujii T, Taguchi H, Katano J, Mori S, et al. Seroprevalence of human herpesvirus 8 in human immunodeficiency virus 1-positive and human immunodeficiency virus 1-negative populations in Japan. J Med Virol 1999;57:159–62. [DOI] [PubMed] [Google Scholar]
- 21.Katano H, Hoshino Y, Morishita Y, et al. Establishing and characterizing a CD 30-positive cell line harboring HHV-8 from a primary effusion lymphoma. J Med Virol 1999;58:394–401. [PubMed] [Google Scholar]
- 22.Miyagi J, Masuda M, Takasu N, et al. Establishment of a primary effusion lymphoma cell line (RM-P1) and in vivo growth system using SCID mice. Int J Hematol 2002;76:165–72. [DOI] [PubMed] [Google Scholar]
- 23.Soulier J, Grollet L, Oksenhendler E, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 1995;86:1276–80. [PubMed] [Google Scholar]
- 24.Gómez-Román JJ, Ocejo-Vinyals G, Sánchez-Velasio P, et al. Presence of human herpesvirus 8 DNA sequences and over-expression of human IL-6 and cyclin D1 in inflammatory myofibroblastic tumor (inflammatory pseudotumor). Lab Invest 2000;80:1121–6. [DOI] [PubMed] [Google Scholar]
- 25.Klussmann JP, Müller A, Wanger M, et al. Human herpesvirus type 8 in salivary gland tumours. J Clin Virol 2000;16:239–46. [DOI] [PubMed] [Google Scholar]
- 26.Koelle DM, Huang M - L, Chandran B, et al. Frequent detection of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) DNA in saliva of human immunodeficiency virus-infected men: clinical and immunologic correlates. J Infect Dis 1997;176:94–102. [DOI] [PubMed] [Google Scholar]
- 27.Lasota J, Miettinen M. Absence of Kaposi’s sarcoma-associated virus (human herpesvirus-8) sequences in angiosarcoma. Virchows Arch 1999;434:51–6. [DOI] [PubMed] [Google Scholar]
- 28.McDonagh DP, Liu J, Gaffey MJ, et al. Detection of Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in angiosarcoma. Am J Pathol 1996;149:1363–8. [PMC free article] [PubMed] [Google Scholar]
- 29.Boralevi F, Masquelier B, Denayrolles M, et al. Study of human herpesvirus 8 (HHV 8) variants from Kaposi’s sarcoma in France: is HHV-8 subtype a responsible for more aggressive tumors? J Infect Dis 1998;178:1546–7. [DOI] [PubMed] [Google Scholar]
- 30.Reed JA, Nador RG, Spaulding D, et al. Demonstration of Kaposi’s sarcoma-associated herpes virus cyclin D homolog in cutaneous Kaposi’s sarcoma by colorimetric in situ hybridization using a catalyzed signal amplification system. Blood 1998;91:3825–32. [PubMed] [Google Scholar]
- 31.Saiki RK, Scharf S, Faloona F, et al. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985;230:1350–4. [DOI] [PubMed] [Google Scholar]
- 32.Russo JJ, Bohenzky R, Chien M-C, et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV 8). Proc Natl Acad Sci U S A 1996;93:14862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchuk D, Drumm M, Saulino A, et al. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res 1990;19:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi’s sarcoma-associated herpesvirus. Virology 1997;236:147–54. [DOI] [PubMed] [Google Scholar]
- 35.Dilnur P, Katano H, Wang ZH, et al. Classic type of Kaposi’s sarcoma and human herpesvirus 8 infection in Xinjiang, China. Pathol Int 2001;51:845–52. [DOI] [PubMed] [Google Scholar]
- 36.Katano H, Sato Y, Kurata T, et al. High expression of HHV-8-encoded ORF73 protein in spindle cells of Kaposi’s sarcoma. Am J Pathol 1999;155:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]