Abstract
Background: In sheep, susceptibility to scrapie, which is similar to human prion diseases such as Kuru and variant Creutzfeldt-Jakob disease (vCJD), is determined by prion protein (PrP) gene (Prnp) polymorphisms. Sheep with genotype ARQ/ARQ, denoting polymorphisms at codons 136, 154, and 171, are susceptible, whereas those with genotypes ARR/ARQ and ARR/ARR are resistant, indicating dominance of ARR over the ARQ allele.
Aims: Based on familial CJD E200K, 129V, where preferential use of the 200E allele in EK heterozygous individuals confers resistance, heterozygous ARR/ARQ sheep were used to test the hypothesis that resistance is caused by preferential use of the ARR allele.
Methods: After assessment of equivalent PrP expression across genotypes, allele use was analysed by sequencing reverse transcription polymerase chain reaction derived DNA clones containing the Prnp gene coding sequence.
Results: The ARR to ARQ ratio was 1.1 in 133 clones, representing Prnp mRNA from three ARR/ARQ sheep, indicating equal use of both alleles.
Conclusions: Dominance of the resistant associated allele in sheep scrapie involves mechanisms other than the absence of PrP derived from the disease associated ARQ allele.
Keywords: genetic resistance, prions, scrapie, transmissible spongiform encephalopathy
Scrapie is a prion disease in sheep that, similar to Kuru or variant Creutzfeldt-Jakob disease (vCJD), requires both prion exposure and a genetic susceptibility. Susceptibility is defined by amino acid polymorphisms in the single copy prion protein (PrP) gene, Prnp. 1 Specifically, in sheep homozygous for A at position 136 and R at position 154, susceptibility is defined by R or Q at position 171. 2 The genotype ARQ/ARQ confers susceptibility, whereas (apart from a few reported exceptions) genotypes ARR/ARQ and ARR/ARR confer resistance. Similarly, a M/V polymorphism at Prnp codon 129 determines susceptibility to vCJD 3 and Kuru, 4 with partial resistance conferred by the V allele. Assuming that overall PrP expression is not affected by genotype, two main alternatives for the resistance of heterozygous individuals exist: PrP ARR is preferentially expressed or PrP ARR interferes with the conversion of PrP ARQ to PrPsc.
There is a precedence for both protein–protein interference and preferential allelic expression in genetic resistance to acquired and hereditary transmissible spongiform encephalopathies (TSEs), respectively. In the case of protein–protein interference, dominant negative inhibition can be defined as the ability of PrP expressed from a resistance associated allele to render conversion of PrP to PrPsc inefficient. It has been shown that C-terminal residues in PrP determine the ability of PrPc to convert to PrPsc on interaction with exogenous PrPsc, 5 and it has been hypothesised that the affinity of PrPc to an endogenous cofactor (“protein X”) required for conversion accounts for this phenomenon. 5, 6 A putative binding region for protein X has been identified. 7
“In the case of protein–protein interference, dominant negative inhibition can be defined as the ability of prion protein (PrP) expressed from a resistance associated allele to render conversion of PrP to PrPsc inefficient”
Supporting this concept, similar observations have been made in vivo using a transgenic mouse model. 8 Notably, with regard to sheep scrapie, this last study used Prnp−/− and Prnp+/+ mice transgenic for mouse PrP with a Q167R mutation (corresponding to the Q/R polymorphism at position 171 of sheep PrP). Because the 167R allele rendered Prnp+/+ mice partially resistant to intracerebral inoculation with Rocky Mountain Laboratory prions, it was concluded that the resistance associated 167R allele is dominant over the wild-type allele.
From that study, it follows that overexpression of a resistance associated allele, such as the ARR allele in sheep, could render heterozygous carriers resistant. Indeed, incomplete penetrance and delayed onset of disease in hereditary CJD E200K, 129V have been associated with preferential use of the wild-type 200E allele over the mutant 200K allele for PrP expression in 200EK heterozygous carriers. In five of seven heterozygous individuals with preferential transcription, the E to K ratio was > 50, indicating a near monoallelic origin of PrP in these individuals. 9
METHODS
Because resistance mechanisms might be shared between hereditary and acquired prion diseases, we tested the hypothesis that the ARR allele is transcribed to the exclusion of ARQ in scrapie resistant ARR/ARQ heterozygous sheep. Sites of preclinical, rather than terminal, PrPsc accumulation were chosen for analysis because (a) primary lymphoid PrPsc accumulation is a feature of Kuru-type prion diseases and is essential for natural sheep scrapie 3, 10 ; (b) follicular dendritic reticulum cells (FDCs) at peripheral accumulation sites are early targets of accumulation in sheep scrapie 10 ; and (c) B cells, rather than brain, were used to demonstrate allelic preference in familial CJD E200K. 9 Before assessment of PrP mRNA, sites of preclinical PrPsc accumulation were analysed for PrP expression by the indirect immunofluorescence assay (IFA) and western blotting (WB) in the ARR/ARQ (resistant) and ARQ/ARQ (susceptible) genotypes to ensure overall equivalent PrP expression within and between genotypes. Three juvenile, healthy, not scrapie exposed ARR/ARQ Suffolk lambs (61, 66, and 76) and one matched healthy ARQ/ARQ control lamb (7828) were used. For WB, 100 mg samples of retropharyngeal lymph node were lysed in 900 μl lysis buffer (10mM Tris/HCl (pH 7.5) containing 0.5% NP-40 and 0.5% deoxycholate), incubated with 200 μg/ml DNase, and centrifuged to remove cell debris. The PrP fraction was concentrated by centrifugation of lysate supernatants in a filter device with a 10 kDa cut off. Serial twofold dilutions of concentrated lysate supernatants (20 μl/lane; wet tissue equivalents: 5000, 2500, 1250, 625, and 313 μg) were used in sodium dodecyl sulfate polyacrylamide gel electrophoresis on 14% gels. The positive control comprised 25 ng/lane of ovine recombinant PrP. The anti-PrP monoclonal antibody F99/97.6.1, 11 followed by enhanced chemoluminescence (ECL; Amersham Biosciences, Piscataway, New Jersey, USA), were used for the detection of bands. For IFA, cryostatic sections were obtained from samples of ileum and mesenteric lymph node, fixed in P-fix (methanol/phosphate buffered saline/glacial acetic acid, 50/49/1 vol/vol), and probed with monoclonal antibodies 7A12 12 for PrP and CNA.42 13 for FDCs. PrP immunoreactivity was visualised using isotype specific biotinylated Fabs with subsequent tyramide amplification of an Alexa green signal (Molecular Probes, Eugene, Oregon, USA). FDCs were visualised using antibody Fab fragments (Fabs) conjugated with Texas red (Jackson ImmunoResearch Laboratories, West Grove, Pennsylvania, USA).
RESULTS
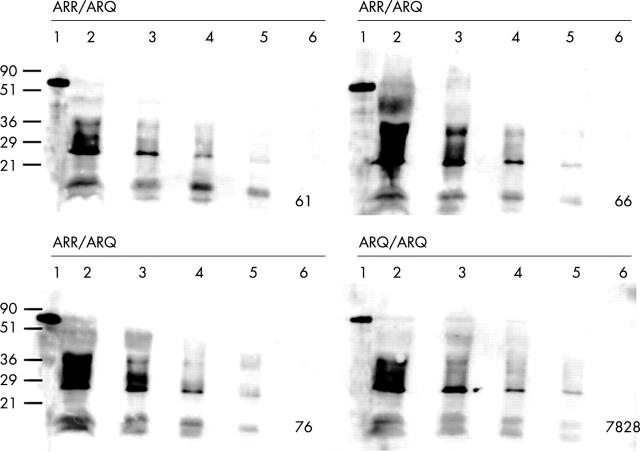
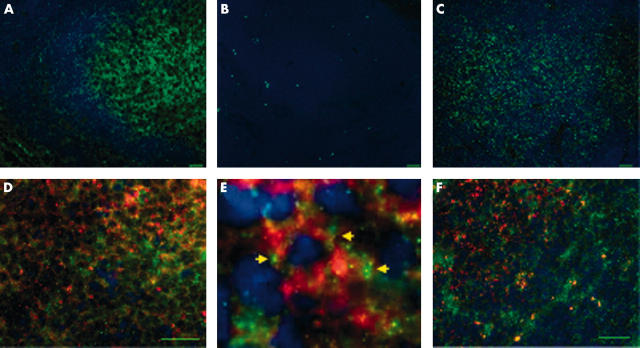
PrP expression was seen in the germinal centre of lymphoid follicles by single label IFA. Genotype specific differences in PrP expression were not evident by the detection limit in WB using serially diluted lymph node lysates (fig 1 ). The PrPc detection limit in two separate experiments was 625 μg wet tissue equivalents for all sheep tested, indicating relative equivalence of PrP expression. Furthermore, PrP signal intensity and distribution detected by IFA were similar for the ARR/ARQ (resistant) and ARQ/ARQ (susceptible) genotypes (fig 2A–C ). Similarly, differences in the cellular specificity of PrP expression by dual IFA (fig 2D–F ) were not found, indicating that genotype specific variations in PrP expression did not confound the interpretation of the allelic ratio of the PrP transcripts. In both genotypes, double label IFA confirmed the expression of PrP in FDCs, cells that are crucial in the pathogenesis of scrapie.
Figure 1.
The detection limit of PrPc by western blotting is identical in three resistant ARR/ARQ sheep (61, 66, 76) and one susceptible ARQ/ARQ control (7828). Twofold dilution series of retropharyngeal lymph node lysates (lane 2, 5000 μg; lane 3, 2500 μg; lane 4, 1250 μg; lane 5, 625 μg; lane 6, 313 μg) were probed with the PrP specific monoclonal antibody F99/97.6.1. 11 Recombinant ovine PrP served as a positive control (lane 1). Molecular masses in kDa are indicated on the left. PrP, prion protein.
Figure 2.
Equivalent prion protein (PrP) expression in gut associated lymphoid tissue from scrapie resistant and susceptible sheep by indirect immunofluorescence assay. (A) Ileal Peyer’s patch from sheep 61 with the resistant PrP genotype ARR/ARQ. PrP is ubiquitously expressed in round cells and stroma of a lymphoid follicle, and staining is particularly strong within the germinal centre. PrP (monoclonal antibody 7A12) is green (Alexa green); nuclei are blue (DAPI); bar, 20 μm. (B) Ileal Peyer’s patch from sheep 61 with resistant PrP genotype ARR/ARQ. No fluorescent signal with irrelevant isotype matched control antibody (anti-Neospora caninum monoclonal antibody 5B6–25); bar, 20 μm. (C) Ileal Peyer’s patch from sheep 7828 with susceptible PrP genotype ARQ/ARQ. Expression of PrP is similar to the heterozygous ARR/ARQ sheep shown in (A). PrP (monoclonal antibody 7A12) is green (Alexa green); nuclei are blue (DAPI); bar, 20 μm. (D) Ileal Peyer’s patch from sheep 66 (ARR/ARQ). Abundant PrP (green) is expressed in follicular dendtritic reticulin cells (FDCs) (red). Dual label immunofluorescence assay (IFA): PrP (7A12), Alexa green; FDC (CNA.42), Texas red; nuclei are blue (DAPI); bar, 20 μm. (E) Detail of panel (D), granular, mostly membrane associated PrP expression (arrows) in an FDC. (F) Ileal Peyer’s patch from sheep 7828 (ARQ/ARQ). Abundant PrP (green) is expressed by FDCs (red) in a pattern indistinguishable from ARR/ARQ sheep. Dual label IFA: PrP (7A12), Alexa green; FDC (CNA.42), Texas red; nuclei are blue (DAPI); bar, 20 μm.
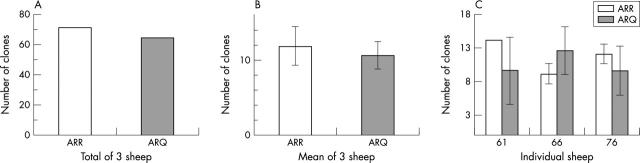
The allelic ratio of PrP transcripts was determined in three ARR/ARQ sheep by sequence analysis of reverse transcription polymerase chain reaction (RT-PCR) derived DNA clones. Cloning and sequencing were chosen over probe based approaches to detect the single base polymorphism at position 171 because the procedures used (RNA isolation, reverse transcription, PCR, and cloning) are random with respect to the single base polymorphism. PrP transcripts isolated from mesenteric (first experiment) or retropharyngeal lymph nodes (second experiment) were analysed in two independent experiments. Total RNA was isolated from lymph nodes and reverse transcribed using oligo-dT primers after DNase digestion. cDNA was amplified by PCR (annealing at 60°C) using the primers PrP148F 5′-AACCGCTATCCACCTCAG and PrP637R 5′-CCACTCGCTCCATTATCT, which span a 491 bp fragment within the PrP coding region containing codons 136, 154, and 171. Absence of genomic DNA in PCR was controlled for by mock RT reactions. PCR products were A–U cloned into pDrive (Qiagen, Valencia, California, USA) and sequenced through a commercial service (Amplicon Express, Pullman, Washington, USA). A single G/A polymorphism at base 365 of the insert corresponding to the second base in codon 171 was detected, as reported previously. 1 The first experiment yielded 60 clones with an R to Q ratio of 37 : 23. The second experiment resulted in 73 clones with a ratio of 33 : 40. Overall, a slight predominance of ARR over ARQ was seen with a ratio of 1.1 (fig 3A ), which was not significantly different from the expected ratio of 1.0 (sign test, p = 0.63). Furthermore, the combined mean number of ARR and ARQ clones of all three sheep did not differ significantly (fig 3B ), and there were no differences between the ARR and ARQ transcripts in the individual sheep (fig 3C ). Accordingly, the hypothesis of differential allelic use, particularly preferential use of 171R, was rejected.
Figure 3.
Use of the ARR and ARQ alleles in cDNA clones of the prion protein gene (Prnp) obtained from the lymph nodes of three ARR/ARQ heterozygous sheep (two experiments). (A) No significant difference in the use of the ARR and ARQ alleles in the total number of Prnp clones examined in all three sheep (sign test, p = 0.63). (B) No significant difference in the use of the ARR and ARQ alleles in the mean of the Prnp clones examined in all three sheep. Error bars are one standard deviation of the mean. (C) No significant difference in the use of the ARR and ARQ alleles in the Prnp clones examined from the individual sheep. Error bars are one standard deviation of two experiments.
Take home messages .
Unlike resistance in hereditary familial Creutzfeldt-Jakob disease E200K, dominance of the resistant associated allele in sheep scrapie involves mechanisms other than the absence of prion protein derived from the disease associated ARQ allele
Mechanisms of genetic resistance vary among transmissible spongiform encephalopathies and may not be shared between hereditary familial and Kuru-type acquired prion diseases
DISCUSSION
Therefore, unlike resistance in hereditary familial CJD E200K, 9 resistance to scrapie in ARR/ARQ heterozygous sheep cannot be attributed to monoallelic mRNA transcription. Rather, our data show that both alleles are used with approximately equal frequency for Prnp transcription. We conclude that mechanisms of genetic resistance vary among TSEs and may not be shared between hereditary familial and Kuru-type acquired prion diseases. The relative genetic resistance of ARR/ARQ sheep must involve mechanisms other than simple absence of PrP ARQ. Such mechanisms may include interference of PrP ARR with PrPsc formation, directly by reducing the likelihood of productive initial interaction with the infectious prion, or indirectly by inhibiting interaction of nascent PrPsc with accessory molecules (protein X hypothesis). 7 Finally, the relative lack of easily convertible substrate PrP ARQ may prolong the incubation period beyond the lifespan of the animal.
“The relative genetic resistance of ARR/ARQ sheep must involve mechanisms other than simple absence of prion protein ARQ”
Acknowledgments
We thank Drs M-S Sy and L Herrmann for the gift of the PrP monoclonal antibody 7A12, D Zhuang for technical assistance, and Dr P Cheevers for discussion of the manuscript. This work was supported by NRICGP/USDA grant 2002-35205-11648. P Caplazi was the recipient of a stipend from the Swiss National Science Foundation, which is gratefully acknowledged.
Abbreviations
CJD, Creutzfeldt-Jakob disease
Fab, antibody Fab fragment(s)
FDC, follicular dendritic reticulum cell
IFA, indirect immunofluorescence assay
PCR, polymerase chain reaction
PrP, prion protein
Prnp, prion protein gene
RT, reverse transcription
TSE, transmissible spongiform encephalopathy
vCJD, Creutzfeldt-Jakob disease, new variant
WB, western blotting
REFERENCES
- 1. Goldmann W, Hunter N, Foster JD, et al. Two alleles of a neural protein gene linked to scrapie in sheep. Proc Natl Acad Sci U S A 1990;87:2476–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O’Rourke KI, Holyoak GR, Clark WW, et al. PrP genotypes and experimental scrapie in orally inoculated Suffolk sheep in the United States. J Gen Virol 1997;78:975–8. [DOI] [PubMed] [Google Scholar]
- 3. Ironside JW. Pathology of variant Creutzfeldt-Jakob disease. Arch Virol Suppl 2000;16:143–51. [DOI] [PubMed] [Google Scholar]
- 4. Cervenakova L, Goldfarb LG, Garruto R, et al. Phenotype–genotype studies in kuru: implications for new variant Creutzfeldt-Jakob disease. Proc Natl Acad Sci U S A 1998;95:13239–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Telling GC, Scott M, Mastrianni J, et al. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell 1995;83:79–90. [DOI] [PubMed] [Google Scholar]
- 6. Zulianello L, Kaneko K, Scott M, et al. Dominant-negative inhibition of prion formation diminished by deletion mutagenesis of the prion protein. J Virol 2000;74:4351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaneko K, Zulianello L, Scott M, et al. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc Natl Acad Sci U S A 1997;94:10069–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perrier V, Kaneko K, Safar J, et al. Dominant-negative inhibition of prion replication in transgenic mice. Proc Natl Acad Sci U S A 2002;99:13079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosenmann H, Halimi M, Kahana I, et al. Differential allelic expression of PrP mRNA in carriers of the E200K mutation. Neurology 1997;49:851–6. [DOI] [PubMed] [Google Scholar]
- 10. Heggebo R, Press CM, Gunnes G, et al. Distribution and accumulation of PrP in gut-associated and peripheral lymphoid tissue of scrapie-affected Suffolk sheep. J Gen Virol 2002;83:479–89. [DOI] [PubMed] [Google Scholar]
- 11. O’Rourke KI, Baszler TV, Besser TE, et al. Preclinical diagnosis of scrapie by immunohistochemistry of third eyelid lymphoid tissue. J Clin Microbiol 2000;38:3254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li R, Liu T, Wong BS, et al. Identification of an epitope in the C terminus of normal prion protein whose expression is modulated by binding events in the N terminus. J Mol Biol 2000;301:567–73. [DOI] [PubMed] [Google Scholar]
- 13. Raymond I, Al Saati T, Tkaczuk J, et al. CNA.42, a new monoclonal antibody directed against a fixative-resistant antigen of follicular dendritic reticulum cells. Am J Pathol 1997;151:1577–85. [PMC free article] [PubMed] [Google Scholar]