Abstract
Aims: To carry out a retrospective study, screening for mutations of the entire coding region of RB1 and adjacent intronic regions in patients with retinoblastoma.
Methods: Mutation screening in DNA extracts of formalin fixed, paraffin wax embedded tissues of 28 patients using combined “exon by exon” polymerase chain reaction mediated single strand conformational polymorphism analysis, followed by DNA sequencing.
Results: Eleven mutations were found in 10 patients. Ten mutations consisted of single base substitutions; 10 were localised in exonic regions (eight nonsense, one missense, and one frameshift) and another one in the intron–exon splicing region. Three novel mutations were identified: a 2 bp insertion in exon 2 (g.5506–5507insAG, R73fsX77), a G to A transition affecting the last invariant nucleotide of intron 13 (g.76429G>A), and a T to C transition in exon 20 (g.156795T>C, L688P). In addition, eight C to T transitions, resulting in stop codons, were found in five different CGA codons (g.64348C>T, g.76430C>T, g.78238C>T, g.78250C>T, and g.150037C>T). Although specific mutation hotspots have not been identified in the literature, eight of the 11 mutations occurred in CGA codons and seven fell within the E1A binding domains (codons 393–572 and 646–772), whereas five were of both types—in CGA codons within E1A binding domains.
Conclusions: CGA codons and E1A binding domains are apparently more frequent mutational targets and should be initially screened in patients with retinoblastoma. Paraffin wax embedded samples proved to be valuable sources of DNA for retrospective studies, providing useful information for genetic counselling.
Keywords: Brazil, genetic counselling, mutational screening, paraffin wax embedded tissues, retinoblastoma, RB1, single strand conformational polymorphism
Retinoblastoma (RB1; OMIM 180200) is the most common intraocular tumour in children, mainly affecting children under 4 years of age, with a prevalence of 1/12 000 to 1/20 000 live births. 1 The development of this malignancy, either in the familial or sporadic form, requires both alleles of the tumour suppressor gene RB1 (GenBank accession number L11910) to be functionless. 2 The familial form accounts for 20–30% of all cases and both eyes are generally affected. In this form, the first mutational event is inherited and the tumour phenotype segregates as an autosomal dominant trait, with 90% penetrance. 3 Conversely, in the sporadic form, the first mutational event might be inherited as a familial trait or as a de novo germ cell mutation in affected children with bilateral retinoblastoma and in 15% of unilateral cases. In contrast, the first mutational event is somatically acquired in the remaining 85% of unilaterally affected patients and in all cases, the second mutational event is somatic. Patients with a constitutional mutation exhibit a higher frequency of secondary tumours in adult life. 4
A wide spectrum of RB1 alterations has been associated with retinoblastoma, including single base substitutions in exonic regions 5– 14 and intron–exon junctions, 5, 9, 10, 12, 13, 15, 16 small deletions/insertions, 17, 18 large deletions, 19, 20 and CpG island hypermethylation at the promoter region. 21– 23
“Patients with a constitutional mutation exhibit a higher frequency of secondary tumours in adult life”
Here, we report RB1 mutations in DNA samples isolated from formalin fixed, paraffin wax embedded tumour tissues in a group of 28 patients with retinoblastoma. The whole coding region (27 exons) and nearby intronic regions were initially screened by polymerase chain reaction (PCR) mediated single strand conformational polymorphism (SSCP) analysis and subsequently by direct DNA sequencing.
MATERIAL AND METHODS
Patients and DNA extraction
We carried out a retrospective study in 25 sporadic cases (13 bilateral and 12 unilateral) and three patients with familial retinoblastoma from the Instituto Nacional de Câncer, Brazil. Paraffin wax blocks were selected after the analysis of histological preparations. In patients with bilateral malignancy, samples from both eyes, when available, were analysed independently. DNA was isolated from paraffin wax as described previously. 24 Different DNA concentrations were used in PCR amplifications based on DNA integrity.
After the identification of mutations in tissue samples, DNA extracted from blood was used for confirming or excluding the constitutional origin of mutations in seven patients (HC1, HC7, HC9, HC17, HC19, HC27, and HC38). In addition, blood samples were obtained from both parents of patients HC1, HC9, and HC27. Two other patients (HC3 and HC22) were not alive at the time of our study and another one (HC6) could not be contacted. DNA from normal samples was used as negative controls. The isolation of DNA from peripheral blood was carried out by standard procedures. 25
PCR amplifications
Exons 1–26 and the coding region of exon 27 were amplified in separate PCR assays, except for exons 15–16, which were amplified in the same reaction. Table 1 shows the primer sequences, positions, and expected fragment sizes of the amplified products.
Table 1.
Primer sequences, fragment sizes in base pairs (bp), and positions in RB1
| Primer | Sequence (5′→3′) | Expected fragment size in bp and RB1 position | Reference |
| 1 F | CTCCCCGGCGCTCCTCCACAGC | 231 (2010–2240) | 1 |
| 1 R | GGGCGCCCTTCCCGCGTGAG | 2 | |
| 2 F | AACAAGTATGTACTGAATCAATTTG | 281 (5355–5635) | 2 |
| 2 R | CTATCTTTCAATTTTTGTATAGTGA | 1 | |
| 3 F | GCGAATTCGGAGGTTATATTCAAAAGAA | 116 (39446–39561) | 3 |
| 3 R | GCGAATTCTGATTTCGTATGTTTTTC | 3 | |
| 4 F | ACAAATTTTTAAGGTTACTGATTTAC | 237 (41876–42112) | 2 |
| 4 R | CCAGAATCTAATTGTGAACAATGAC | 2 | |
| 5 F | AACTACTATGACTTCTAAATTACG | 221 (44607–44827) | 2 |
| 5 R | CTTAATTTATGAAGTAGCCTGCTA | 1 | |
| 6 F | CTGGAAAACTTTCTTTCAGTGATA | 210 (45734–45943) | 1 |
| 6 R | GGAATTTAGTCCAAAGGAATGCC | 2 | |
| 7 F | ATACAAAGATCTGAATCTCTAACT | 226 (56800–57025) | 1 |
| 7 R | CTAGACATTCAATAAGCAACTGC | 1 | |
| 8 F | GCGAATTCAAACAGCTGTTATACCCATT | 143 (59651–59793) | 3 |
| 8 R | GCGAATTCTCATCTATATTACATTCAT | 3 | |
| 9 F | GTTCAAGAGTCAAGAGATTAGATT | 209 (61661–61869) | 1 |
| 9 R | CAATTATCCTCCCTCCACAGTCTCA | 1 | |
| 10 F | GCCTCTGTGTGCTGAGAGATGTA | 277 (64264–64540) | 2 |
| 10 R | AATGATATCTAAAGGTCACTAAGC | 1 | |
| 11 F | GATTTTATGAGACAACAGAAGCA | 244 (65264–65507) | 1 |
| 11 R | ATCTGAAACACTATAAAGCCATG | 1 | |
| 12 F | AGAGACAAGTGGGAGGCAGTG | 327 (70120–70446) | 1 |
| 12 R | GATAACTACATGTTAGATAGGAG | 1 | |
| 13 F | CTGATTACACAGTATCCTCGAC | 239 (73704–73942) | 2 |
| 13 R | ATACGAACTGGAAAGATGCTGC | 2 | |
| 14 F | ATTGTGATTTTCTAAAATAGCAGG | 229 (76377–76605) | 1 |
| 14 R | CAGGATGATCTTGATGCCTTG | 1 | |
| 15–16 F | CAATGCTGACACAAATAAGGTT | 321 (76822–77142) | 1 |
| 15–16 R | AAGAAACACACCACATTTTAACT | 1 | |
| 17 F | AGCTCAAGGGTTAATATTTCATAA | 302 (78032–78333) | 1 |
| 17 R | AATTTGTTAGCCATATGCACATG | 1 | |
| 18 F | ATGTACCTGGGAAAATTATGCTT | 258 (149939–150196) | 1 |
| 18 R | CTTTATAGAATGTTACATTGCAC | 1 | |
| 19 F | ATCTGTGATTCTTAGCCAACTTG | 250 (153154–153403) | 2 |
| 19 R | AGTCAGCCTAGTTTCAGAGTC | 1 | |
| 20 F | CTGGGGGAAAGAAAAGAGTGG | 328 (156544–156871) | 1 |
| 20 R | GAGGAGAGAAGGTGAAGTGCT | 1 | |
| 21 F | GAACAAAACCATGTAATAAAATTCT | 219 (160676–160894) | 1 |
| 21 R | ACCTATGTTATGTTATGGATATGG | 2 | |
| 22 F | ACTGTTCTTCCTCAGACATTCA | 182 (161982–162163) | 1 |
| 22 R | TTGGTGGACCCATTACATTAGA | 1 | |
| 23 F | CTAATGTAATGGGTCCACCAAA | 270 (162143–162412) | 1 |
| 23 R | TCCCCCTCTCATTCTTTACTAC | 1 | |
| 24 F | TCATCTCTGCAAAATTGTATATGG | 217 (170309–170525) | 1 |
| 24 R | TATGCAATATGCCTGGATGAGG | 1 | |
| 25 F | TTGCTAACTATGAAACACTGGC | 247 (173647–173893) | 1 |
| 25 R | ATGACCATCTCAGCTACTGGA | 1 | |
| 26 F | TCGAAAGCATCATAGTTACTGG | 248 (174279–174526) | 1 |
| 26 R | ATGCATAAACAAACCTGCCAACT | 1 | |
| 27 F | TGCAAGGTCCTGAGCGCCAT | 238 (176894–177131) | 1 |
| 27 R | GAGAGACAATGAATCCAGAGGTG | 1 |
PCR amplifications were performed in 50 μl mixes containing 20mM Tris, 50μM KCl, 1.5mM MgCl2, 50μM of each dNTP, 20 pmol of each primer, 1 U of Taq DNA polymerase, and variable amounts (1–5 μl) of paraffin wax extracted DNA. Amplification conditions consisted of one initial denaturing step of two minutes at 94°C, followed by 40 cycles of one minute at 94°C, 35 seconds at 52°C (55°C for exons 2, 3, 10 and 13, and 65°C for exon 1), and 40 seconds at 72°C, with a final extension step of four minutes at 72°C.
SSCP analyses
To increase the sensitivity of SSCP, all PCR products were screened using two different, non-denaturing gel conditions. Gels were prepared with 0.5× MDE gel solution (FMC Bioproducts, Rockland, New York, USA), with or without 6% (vol/vol) glycerol (dissolved in 0.5× TBE buffer). Aliquots of amplified products were mixed with equal amounts of denaturing buffer (50% deionised formamide and 50% glycerol with 0.05% xylene cyanol and 0.05% bromophenol blue). Samples were denatured at 95°C for five minutes, kept on ice until loading on to gels, and run in vertical electrophoretic plates (model V16; Gibco BRL, Gaithersburgh, Maryland, USA) at constant voltage (between 3 and 5 V/cm) for 15–18 hours at room temperature. SSCP fragments were visualised by silver staining. Amplification products showing abnormal SSCP patterns were subsequently sequenced. In addition, all 27 exons of six patients with sporadic retinoblastoma who showed normal SSCP patterns were also sequenced (unilaterally affected HC6, HC11, and HC14 and bilaterally affected HC4, HC27, and HC37).
DNA sequencing
PCR products were purified with the GFX™ PCR DNA and Gel Band Purification Kit® (Pharmacia, Uppsala, Sweden). They were subsequently labelled with the DYEnamyc™ ET Terminator Cycle Sequencing Premix Kit® (Pharmacia). Products were electrophoresed in an ABI PRISM™ 377 automatic sequencer (Applied Biosystems, Foster City, California, USA) and analysed with Sequence Navigator software (Perkin Elmer, Foster City, California, USA). Novel mutations were deposited in GenBank (RB1–OMIM: 180200; GDB: 118734; GenBank: L11910; HGMD: RB1; http://www.d-lohmann.de/Rb/mutations.html (RB1 gene mutation database); see table 2 for accession numbers).
Table 2.
List of mutations identified in our study
| Patient | Sex | Tumour form/type | Mutation | Effect | Localisation | Method of detection | GenBank number |
| HC17 | Male | S/B | g.5506-5507insAG | R73fsX77 | E 2 | SEQ | AY282604 |
| g.78238C>T | R552X | E 17 | SSCP 2 | L41924 | |||
| HC6 | Male | S/U | g.64348C>T | R320X | E 10 | SEQ | AF532992 |
| HC1 | Male | S/U | g.76429G>A | Abnormal splicing | I 13 | SSCP 1 | AY124935 |
| HC27 | Female | S/B | g.76430C>T | R445X | E 14 | SEQ | AF532990 |
| HC7 | Female | F/B | g.78238C>T | R552X | E 17 | SSCP 2 | L41924 |
| HC38 | Female | F/B | g.78238C>T | R552X | E 17 | SSCP 2 | L41924 |
| HC9 | Male | S/B | g.78250C>T | R556X | E 17 | SSCP 2 | L41904 |
| HC3 | Male | S/B | g.150037C>T | R579X | E 18 | SSCP 1, 2 | AF532991 |
| HC19 | Female | S/U | g.150037C>T | R579X | E 18 | SSCP 1, 2 | AF532991 |
| HC22 | Male | S/U | g.156795T>C | L688P | E 20 | SSCP 2 | AY124936 |
Novel mutations are shown in bold and constitutional mutations are underlined. The mutation nomenclature was based in Dunnen and Antonarakis. 26 Nucleotide positions and codon identification numbers are based on the normal RB1 sequence (GenBank accession L11910).
B, bilaterally affected patients; E, exon; F, familial; I, intron; S, sporadic; U, unilaterally affected patients; SEQ, direct DNA sequencing; SSCP 1 , single strand conformational polymorphism gel without glycerol; SSCP 2 , single strand conformational polymorphism gel with glycerol.
RESULTS
The SSCP assays were sensitive enough to detect mutations in eight patients, although sensitivity was affected by glycerol because some abnormal patterns were only apparent with or without this additive. In total, 11 mutations were detected in 10 patients (table 2 ), corresponding to one mutation for each patient except for patient HC17, who carried two mutations. Eight different mutations were identified because one (g.78238C>T) was present in three patients and another one (g.150037C>T) was seen in two patients. Seven of these eight mutations were single base substitutions; seven in exonic regions (five nonsense, one missense, and one frameshift) and another one in an intron–exon splicing region. Five mutations consisted of C → T transitions at CGA codons, five fell in the E1A binding domains (codons 393–572 and 646–772), whereas three shared both characteristics.
Three novel mutations were identified. The first one, detected in DNA extracted from the blood of patient HC17, was an AG insertion in exon 2, resulting in a premature stop codon (g.5506–5507insAG, R73fsX77). It was identified exclusively by sequencing after the initial detection of a different mutation (g.78238C>T, R552X in exon 17) in tumour DNA of the patient’s left eye, which was absent in the tumour mass of the right eye or blood DNA. These findings proved that the novel mutation was constitutional and that the one present in the tumour mass originated as a second mutational event. Interestingly, this second mutation was found to be constitutional in patients HC7 and HC38 (see below).
A second novel mutation consisted of a G → A transition affecting the last (3′) nucleotide of intron 13 (g.76429G>A; fig 1 ), at a crucial site that needs to remain invariant for normal splicing in a sporadically, unilaterally affected patient (HC1). This mutation, although constitutional, could not be found when sequencing exon 14 and adjacent regions in blood DNA from either parent, so that it must have been inherited as a de novo germ line mutation. A third novel mutation, consisting of a T → C transition in exon 20 (g.156795T>C, L688P), was detected in tumour tissue of patient HC22 who was dead at the time of our study.
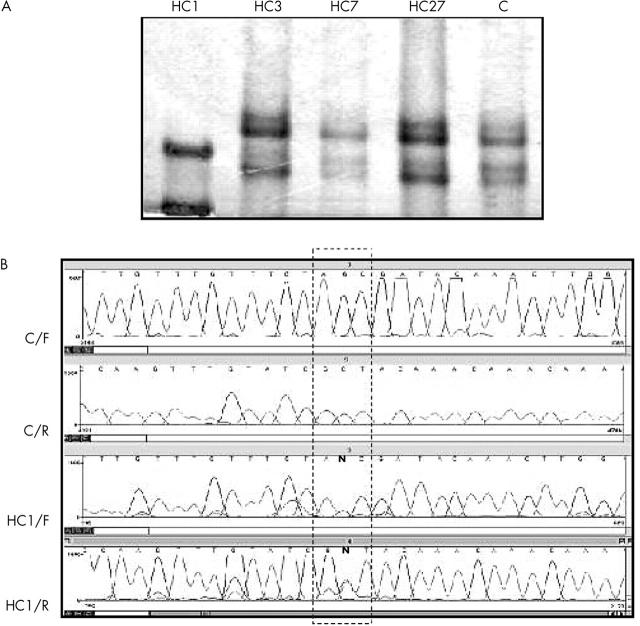
Figure 1.
(A) Single strand conformational polymorphism (SSCP) analyses of exon 14 and adjacent intronic regions. Amplified products of tumour DNA of patient HC1 show a different migration pattern when compared with the normal control (C) and other patients. (B) Comparative DNA sequencing between a control and patient HC1. C/F, normal control with the forward primer; C/R, normal control with the reverse primer; HC1/F, DNA of patient HC1 with the forward primer; HC1/R, DNA of patient HC1 with the reverse primer. In the box, the letter “N” indicates simultaneous presence of the normal base (G and C in forward and reverse strands respectively) and A and T, respectively, in patient HC1. This mutation was seen both in tumour and blood DNA samples, confirming its constitutional origin. Note that patient HC27 shows a normal SSCP pattern, although DNA sequencing identified a transition one base apart (table 2 ).
Five mutations in eight patients consisted of C → T transitions changing different CGA codons to TGA (Arg → Stop). One of them (g.78238C>T) was found in patient HC17 and in the half sisters HC7 and HC38. In the half sisters, this mutation was constitutional. A second mutation, g.64348C>T, was only detected by sequencing tumour DNA of patient HC6, who showed normal SSCP patterns, although its constitutional origin could not be investigated because blood samples were not available. A third mutation, g.76430C>T, was only detected after tumour DNA sequencing in patient HC27, who showed normal SSCP patterns (fig 1A ). This mutation could not be found in either of her parents. A fourth mutation, g.78250C>T, found in separate analyses of both eyes of patient HC9 was shown to be constitutional. The SSCP pattern of the DNA from the right eye differed from the normal control, whereas DNA from the left eye showed a mixed SSCP pattern between the control and DNA from the right eye (fig 2 ). Sequencing confirmed this heterogeneity showing only the mutated base (T) in the right eye and both the mutated (T) and the normal base (C) in the left eye. This mutation was not found in either of his parents. Finally, a fifth mutation, g.150037C>T, was found in tumour tissue of patient HC3, who was not alive at the time of our study, and in patient HC19. Sequencing of exon 18 and adjacent regions in blood DNA in patient HC19 showed that the mutation was not constitutional.
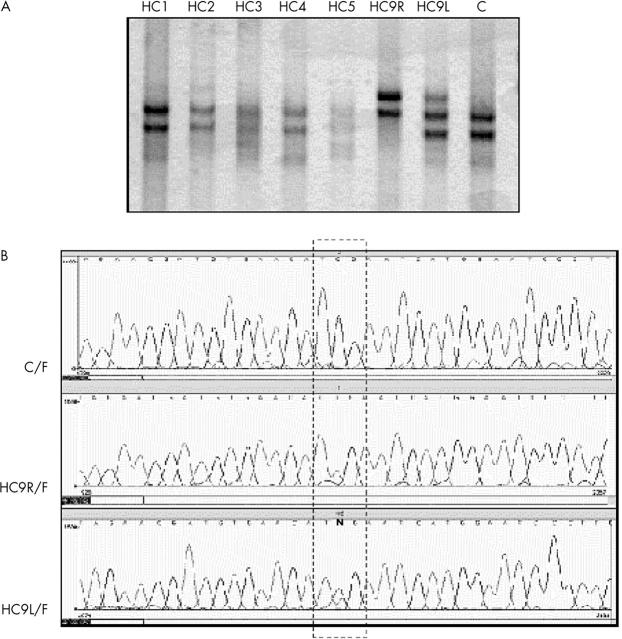
Figure 2.
(A) Single strand conformational polymorphism (SSCP) analyses of exon 17 and adjacent introns. Tumour DNA was obtained from both eyes of patient HC9 and analysed separately (HC9R, DNA from right eye; HC9L, DNA from left eye). HC9R shows a different migration pattern when compared with the normal control (C) and other patients, whereas HC9L shows a mixed pattern between normal and HC9R. (B) Comparative DNA sequencing between control, HC9R, and HC9L. C/F, normal control with the forward primer; HC9R/F, tumour DNA from the right eye with the forward primer; HC9L/F, tumour DNA from the left eye with the forward primer. Sequencing confirmed the SSCP patterns, showing only T in HC9R and the presence of C (normal base) and T (mutated base) in HC9L (indicated by “N”). These data were confirmed using reverse sequencing (not shown).
DISCUSSION
Because tumour DNA was extracted from formalin fixed, paraffin wax embedded blocks only small quantities of degraded DNA were obtained. For this reason, we chose a detection strategy based on screening entire coding regions and adjacent intronic regions using PCR mediated SSCP analysis, followed by direct DNA sequencing. Based on this combined methodology we identified mutations in eight patients. In addition, mutations not detected by SSCP were identified by DNA sequencing in three patients. The mutations described here consisted of 10 single base substitutions and one small insertion.
A constitutional, frameshift mutation was present in patient HC17, comprising a novel AG insertion in exon 2, which resulted in a stop codon at position 77. The presence of direct (AG)5 repeats at the insertion site suggests that slipped mispairing or misalignments are the most probable mechanisms responsible for this mutation. 27 In this patient, a different mutation was detected in the left eye (g.78238C>T, R552X), which was not found in DNA from either the right eye or blood, indicating that it was a second mutational event.
In patient HC1, a novel mutation involved a G → A replacement of the last (3′) base of intron 13; this mutation affected an intronic site (G) previously shown to be strictly invariant in more than 400 vertebrate genes with intronic consensus sequences at 5′ and 3′ junctions. 28 The demonstration of its constitutional origin enabled patient HC1 to be included in the high risk group of unilaterally affected patients with this condition, and the fact that it was absent in his parents indicated that it must have been transmitted as a de novo germline mutation. A third novel mutation, in exon 20, was identified in patient HC22, although its constitutional origin could not be investigated.
“Although specific mutation hotspots have not been identified in the literature, eight of the 11 mutations occurred in CGA codons and seven fell within E1A binding domains”
The remaining mutations consisted of C → T transitions that changed CGA (arginine) codons into TGA (stop) codons, resulting in truncated RB proteins, and that occurred in previously described hotspots. 5, 9– 16, 29, 30 CpG dinucleotides are frequent sites of mutation in a variety of genes including RB1 31, 32 because of frequent methylation of cytosine residues and subsequent deamination resulting in C → T transitions.
The confirmation of the constitutional origin of mutations in patients HC1, HC7, HC9, HC17, HC27, and HC38 allowed their inclusion in a high risk group, in whom careful follow up is recommended because they are more likely to develop secondary tumours in adult life than are non-carriers. Although specific mutation hotspots have not been identified in the literature, eight of the 11 mutations occurred in CGA codons and seven fell within E1A binding domains. Five of these mutations were of both types—that is, they were in CGA codons within E1A binding domains. These sites are apparently more frequent mutational targets and should be initially screened in patients with retinoblastoma.
There are several factors that might have affected our ability to detect mutational events. First, the maximum sensitivity of SSCP is around 80–90%, 25 which is why some mutations cannot be detected, as was the case of patient HC27 who showed a C → T transition at position 76 430, although in patient HC1, abnormal SSCP patterns were seen as a result of a G → A transition occurring only one base apart (fig 1A ). Conversely, SSCP was very sensitive in distinguishing different tumour samples in patient HC9 (fig 2 ), a finding that was later clarified by sequencing. Lack of a normal SSCP pattern indicated loss of heterozygosity in the right eye, which was associated with the second mutational event. Second, in the heterozygous condition, alterations in gene structure, such as entire exonic deletions or mutations affecting priming sites, might be undetected by SSCP or DNA sequencing as a result of amplification of the normal allele. Third, formalin fixed, paraffin wax embedded tumour tissue is not an optimal source of DNA because DNA integrity can be severely affected, precluding the use of other techniques, such as Southern blotting, for detecting large deletions. Finally, DNA may be present at very low concentrations or it may be even absent in extracts of embedded tumour tissues that have undergone severe necrosis. This is especially relevant for patients with unilateral tumours resulting from two independent, somatic mutational events, and could result in underestimates in these patients because the first mutational event occurs in tumour tissue.
Take home messages .
Using polymerase chain reaction mediated single strand conformational polymorphism analysis and subsequent direct DNA sequencing we identified 11 mutations in the RB1 gene in 10 of 28 patients with retinoblastoma
Mutations were found more frequently in CGA codons and E1A binding domains, which should be initially screened in patients with retinoblastoma
Paraffin wax embedded samples are useful sources of DNA for retrospective studies, providing valuable information for genetic counselling
Regardless of these limitations, however, patient HC19 is a good example of successful screening of paraffin wax embedded, tumour samples in which a somatic mutational event was identified, although evidence of a constitutional mutation was lacking. Furthermore, a second mutational event was demonstrated in patient HC17. In addition, paraffin wax embedded samples represent an important source of DNA in cases for retrospective study, and provide valuable information for genetic counselling.
Acknowledgments
This work was supported by Instituto Nacional de Câncer and Fundação Ary Frauzino (Rio de Janeiro, Brazil). E Braggio is a recipient of a scholarship from the Conselho Nacional de Desenvolvimento (Brazil).
Abbreviations
PCR, polymerase chain reaction
SSCP, single strand conformational polymorphism
REFERENCES
- 1. Schultz KR, Ranade S, Neglia JP, et al. An increased relative frequency of retinoblastoma at a rural regional referral hospital in Miraj, Maharashtra, India. Cancer 1993;72:282–6. [DOI] [PubMed] [Google Scholar]
- 2. Knudson AG. Mutations and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A 1971;68:820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vogel F. Genetics of retinoblastoma. Hum Genet 1979;52:1–54. [DOI] [PubMed] [Google Scholar]
- 4. Gallie BL, Moore A. Retinoblastoma. In: Taylor D, ed. Paediatric ophthalmology. Oxford: Blackwell Scientific, 1997:519–53.
- 5. Yandell DW, Campbell TA, Dayton SH, et al. Oncogenic point mutations in the human retinoblastoma gene: their application to genetic counseling. N Engl J Med 1989;321:1689–95. [DOI] [PubMed] [Google Scholar]
- 6. Lohmann DR, Horsthemke B, Gillessen-Kaesbach G, et al. Detection of small RB1 gene deletions in retinoblastoma by multiplex PCR and high-resolution gel electrophoresis. Hum Genet 1992;89:49–53. [DOI] [PubMed] [Google Scholar]
- 7. Lohmann DR, Brandt B, Höpping W, et al. Distinct RB1 gene mutations with low penetrance in hereditary retinoblastoma. Hum Genet 1994a;94:491–6. [DOI] [PubMed] [Google Scholar]
- 8. Lohmann DR, Brandt B, Höpping W, et al. Spectrum of small length germ-line mutations in the RB1 gene. Hum Mol Genet 1994b;3:2187–93. [DOI] [PubMed] [Google Scholar]
- 9. Lohmann DR, Brandt B, Höpping W, et al. The spectrum of RB1 germ-line mutations in hereditary retinoblastoma. Am J Hum Genet 1996;58:940–9. [PMC free article] [PubMed] [Google Scholar]
- 10. Lohmann DR, Gerick M, Brandt B, et al. Constitutional RB1 gene mutations in patients with isolated unilateral retinoblastoma. Am J Hum Genet 1997;61:282–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cowell JK, Smith T, Bia B. Frequent constitutional C to T mutations in CGA-arginine codons in the RB1 gene produce premature stop codons in patients with bilateral (hereditary) retinoblastoma. Eur J Hum Genet 1994;2:281–90. [DOI] [PubMed] [Google Scholar]
- 12. Shimizu T, Toguchida J, Kato MV, et al. Detection of mutations of the RB1 gene in retinoblastoma patients by using exon-by-exon PCR-SSCP analysis. Am J Hum Genet 1994;54:793–800. [PMC free article] [PubMed] [Google Scholar]
- 13. Blanquet V, Turleau C, Gross-Morand MS, et al. Spectrum of germ-line mutations in the RB1 gene: a study of 232 patients with hereditary and non hereditary retinoblastoma. Hum Mol Genet 1995;4:383–8. [DOI] [PubMed] [Google Scholar]
- 14. Liu Z, Song Y, Bia B, et al. Germ-line mutations in the RB1 gene in patients with hereditary retinoblastoma. Genes Chromosomes Cancer 1995;14:277–84. [DOI] [PubMed] [Google Scholar]
- 15. Yilmaz S, Horsthemke B, Lohmann DR. Twelve novel RB1 gene mutations in patients with hereditary retinoblastoma. Hum Mutat 1998;12:434. [DOI] [PubMed] [Google Scholar]
- 16. Klutz M, Horsthemke B, Lohmann DR. RB1 gene mutations in peripheral blood DNA of patients with isolated unilateral retinoblastoma. Am J Hum Genet 1999;64:666–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Canning S, Dryja TP. Short, direct repeats at the breakpoints of deletions of the retinoblastoma gene. Genetics 1989;86:5044–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kloss K, Währisch P, Greger V, et al. Characterization of deletions at the retinoblastoma locus in patients with bilateral retinoblastoma. Am J Med Genet 1991;39:196–200. [DOI] [PubMed] [Google Scholar]
- 19. Squire J, Phillips RA, Boyce S, et al. A detailed analysis of chromosomal changes in heritable and non-heritable retinoblastoma. Hum Genet 1985;70:291–301. [DOI] [PubMed] [Google Scholar]
- 20. Cowell JK, Hogg A. Genetics and cytogenetics of retinoblastoma. Cancer Genet Cytogenet 1992;64:1–11. [DOI] [PubMed] [Google Scholar]
- 21. Sakai T, Ohtani N, McGee TL, et al. Oncogenic germ-line mutations in Sp1 and ATF sites in the human retinoblastoma gene. Nature 1991;353:83–6. [DOI] [PubMed] [Google Scholar]
- 22. Cowell JK, Bia B, Akoulitchev A. A novel mutation in the promoter region in a family with a mild form of retinoblastoma indicates the location of a new regulatory domain for the RB1 gene. Oncogene 1996;12:431–6. [PubMed] [Google Scholar]
- 23. Zajaczek S, Jakubowska A, Kurzawski G, et al. Age at diagnosis to discriminate those patients for whom constitutional DNA sequencing is appropriate in sporadic unilateral retinoblastoma. Eur J Cancer 1998;34:1919–21. [DOI] [PubMed] [Google Scholar]
- 24. Stefanoff CG, Hassan R, Gonzalez AC, et al. Laboratory strategies for efficient handling of paraffin-embedded tissues for molecular detection of clonality in non-Hodgkin lymphomas. Diagn Mol Pathol 2003;12:79–87. [DOI] [PubMed] [Google Scholar]
- 25. Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbour, 2001.
- 26. Dunnen JT, Antorarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 2000;15:7–12. [DOI] [PubMed] [Google Scholar]
- 27. Krawczak M, Cooper DN. Gene deletions causing human genetic disease: mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum Genet 1991;86:425–41. [DOI] [PubMed] [Google Scholar]
- 28. Padgett RA, Grabowski PJ, Konarska MM, et al. Splicing of messenger RNA precursors. Annu Rev Biochem 1986;55:1119–50. [DOI] [PubMed] [Google Scholar]
- 29. Hogg A, Britta B, Onadim Z, et al. Molecular mechanisms of oncogenic mutations in tumors from patients with bilateral and unilateral retinoblastoma. Proc Natl Acad Sci U S A 1993;90:7351–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lohmann DR. RB1 gene mutations in retinoblastoma. Hum Mutat 1999;14:283–8. [DOI] [PubMed] [Google Scholar]
- 31. Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet 1989;83:181–8. [DOI] [PubMed] [Google Scholar]
- 32. Tasheva ES, Roufa DJ. Deoxycytidine methylation and the origin of spontaneous transition mutations in mammalian cells. Somat Cell Mol Genet 1993;19:275–83. [DOI] [PubMed] [Google Scholar]