Abstract
Aim: To examine three lineages of Toxoplasma gondii RH strain in terms of performance in the dye test, culture, and gene expression.
Methods: Historical data (culture growth and performance in the dye test) from three lineages of RH strain tachyzoites (B, J, and Q) that had been continuously cultured in HeLa cells was assessed. Tachyzoite harvests obtained during continuous cell culture were retrieved from liquid nitrogen and cultured in HeLa cells, providing mRNA that was extracted and used to study gene expression using random amplified polymorphic DNA analysis at different stages of lineage adaptation to continuous culture.
Results: The B and Q lineages consistently produced tachyzoites that were successfully used in the dye test and their gene expression was stable after multiple passages. The J lineage had unpredictable growth, tachyzoites unsuitable for use in the dye test, and changing gene expression with multiple passage.
Conclusion: This study has explained some anomalies in the performance of different stocks of T gondii, and suggests that lineages that are still evolving in cell culture should be avoided.
Keywords: random amplified polymorphic DNA analysis, RH strain, Toxoplasma gondii, continuous cell culture, gene expression
Tachyzoites of the non-cyst forming RH strain of type I Toxoplasma gondii are routinely grown in continuous cell culture for use as diagnostic test reagents. 1, 2 Different lineages originate from different stored passes of in vivo derived RH strain so that any lineage differences may result from changes in gene expression with passage. 2 Gene expression changes have been seen in RH strain tachyzoites since 1939. 3 Restriction fragment length polymorphism and random amplified polymorphic DNA (RAPD) analyses have demonstrated genetic variability in different laboratory stocks of the RH strain. 3, 4 Any changes in gene expression that have arisen before or during continuous passage in HeLa cells may have a bearing on diagnostic assays, and it is important that these changes should be identified.
METHODS
Historical data
Data from three lineages of RH strain tachyzoites (B, J, and Q) that had been continuously cultured in HeLa cells were collated. The usefulness of cell culture was assessed on the ability of each lineage to yield viable tachyzoites, and performance of the tachyzoites in the dye test. The dye test, the serological gold standard, relies on complement mediated killing of T gondii tachyzoites to measure the total amount of specific anti-T gondii antibodies. To ensure dye test success a harvest with a minimum of ⩾ 1 × 106 free tachyzoites/ml and ⩾ 90% viability is required. 1 To perform satisfactorily the dye test had to yield the expected titres (± 1 dilution) for control sera, and have a clearly defined end point dilution (50% killing).
Sample preparation
Cell culture
Tachyzoite harvests, approximately every five passes, were stored in liquid nitrogen. Tachyzoites from three pass numbers from each of the above RH strain lineages (B, J, and Q) were rapidly thawed and cultured as described by Chatterton et al. 5 Uninfected HeLa cells were used as a control. Tachyzoites were added to confluent HeLa monolayers in 25 cm3 cell culture flasks (Corning, High Wycome, UK) containing maintenance medium: Eagle’s minimum essential medium with Earle’s balanced salt and 25mM Hepes solution (Biowhittaker, Wokingham, UK), supplemented with 2% fetal bovine serum, 2mM L-glutamine (Biowhittaker), 40 000 U gentamicin (Roussel Laboratories, Uxbridge, UK), and 1 mg Fungizone (Squibb and Sons, Hounslow, UK) and incubated at 37°C with 5% CO2. After six hours the medium was replaced by fresh maintenance medium, then serum free medium after 24 hours. Cultures were examined by microscope on a daily basis for tachyzoite growth and biofilm stability, and passed blind two to three times until > 1.0 × 106 free tachyzoites/ml were produced. The final harvest, containing free tachyzoites and an infected cell monolayer, was centrifuged at 1750 ×g for 10 minutes, and washed once in phosphate buffered saline.
cDNA production and assessment
The tachyzoite/cell pellet was lysed and mRNA extracted on to magnetic mRNA capture beads according to the manufacturer’s instructions (Dynabeads mRNA DIRECT™ micro kit; Dynal AS, Oslo, Norway). 6 cDNA was produced by a two step procedure. First, extracted mRNA was suspended in 25 μl annealing mix (20 μl H2O and 5 μl 5× M-MLV RT reaction buffer; Promega, Southampton, UK), covered with a drop of mineral oil (Sigma Aldrich Co Ltd, Poole, Dorset, UK), heated at 70°C for five minutes, and then cooled on ice. Second, 25 μl reverse transcriptase reaction mix (5 μl 5× M-MLV RT reaction buffer (Promega), 4mM dNTP mix (Advanced Biotechnologies, Leatherhead, Surrey, UK), 25 units ribonuclease inhibitor (Superase-In; Ambion Inc, Abingdon, Cambridgeshire, UK), and 200 units M-MLV RT (Promega)) were added to the RNA, which was then incubated at 42°C for one hour.
The amount of cellular cDNA was measured by polymerase chain reaction (PCR) amplification of the cellular housekeeping hypoxanthine phosphoribosyl transferase (HPRT) gene using the following primers: HPRTa (5′ GAC CGT CAA CAG GGG ACA T 3′) and HPRTb (5′ CGA CCT TGA CCA TCT TTG GA 3′) (Qiagen, Crawley, Surrey, UK). Toxoplasma gondii cDNA was measured by PCR amplification of the conserved T gondii B1 gene using primers S114 (5′ GGAACTGCATCCGTTCATGAG 3′) and S115 (5′ TCTTTAAAGCGTTCGTGGTC 3′) (Severn Biotech, Kidderminster, UK). 7 Briefly, 2.5 μl cDNA was added to 25 μl master mix (0.4μM each primer, 2.5 μl 10× PCR buffer/15mM MgCl2 (Qiagen), and 0.6125 units Hot Start Taq polymerase (Qiagen). HPRT PCR also incorporated 0.4mM dNTP mix (Advanced Biotechnologies) and was carried out as follows: an initial 95°C for 15 minutes, then 35 cycles of 95°C for 45 seconds, 55°C for 90 seconds, and 72°C for 90 seconds, with a final extension step at 72°C for 10 minutes. B1 PCR used 0.8mM dNTP mix (Advanced Biotechnologies) and was carried out at 95°C for 15 minutes, with 40 cycles of 94°C for one minute, 53°C for one minute, and 72°C for one minute, followed by a final extension at 72°C for five minutes. Products were viewed on a 2.5% agarose gel containing ethidium bromide.
RAPD
RAPD uses short, random, single primers in a PCR to amplify multiple segments of the genome, in this case, either T gondii or the host cell. 4 Using primers from a study by Guo et al, 4 RAPD was performed on cDNA to compare specific segments of mRNA expressed during culture. Each assay incorporated one of three primers (B05, 5′TGCGCCCTTC3′; B12, 5′ CCTTGACGCA 3′; or F15, 5′ CCTGTACTCC 3′). 4 Sample cDNA was diluted to ensure that the same amount of total cDNA (determined by quantitative PCR of cellular and toxoplasma cDNA) was added to each reaction. The 25 μl reaction mix contained 2.0 μl cDNA dilution, 0.833μM random primer (MWG Biotech Ltd, Milton Keynes, UK), 2.5 μl 10× buffer/15mM MgCl2 (Qiagen), 0.8mM dNTP mix, and 1.0 unit of Hot Start Taq polymerase (Qiagen). Reactions were covered with mineral oil. Amplification was at 95°C for 15 minutes, which was followed by three cycles of 94°C for five minutes, 36°C for five minutes, and 72°C for five minutes, then 40 cycles of 94°C for one minute, 36°C for one minute, and 72°C for two minutes, and a final 10 minute extension at 72°C (PHC1 Dri-block). Products were electrophoresed on a 1.25% agarose gel with ethidium bromide in Tris borate EDTA (TBE) buffer for two hours at 150 V, then submerged in TBE buffer with ethidium bromide for 30 minutes. Images were viewed using photo editor software (Olympus, London, UK). Products were identified as T gondii specific if present in the tachyzoite cultures but not the uninfected cell control. Only distinct and reproducible bands (present in at least four of six RAPD runs) were taken into account.
RESULTS
Table 1 shows that the three RH strain lineages had varying abilities to produce good quality harvests that could be used successfully in the dye test. The B and Q lineages adapted very quickly to growth in continuous cell culture, producing harvests at fairly predictable intervals. In contrast, the J lineage cultures were unpredictable and tended to infect and destroy the HeLa cell monolayer much more quickly than the other lineages, leading to decreased tachyzoite viability because the tachyzoites adhered to the flask walls and died.
Table 1.
Effect of RH strain lineage on the production of tachyzoites in routine continuous cell culture system
| Lineage/pass numbers (range) | Harvests ⩾1 × 106 tachyzoites/ml, ⩾90% viable (%) | Dye test success (%) |
| B56–100* | 223/306 (72.3%) | 152/181 (84.4%) |
| B101–188 | 783/825 (94.9%) | 217/234 (92.7%) |
| J1–78 | 144/191 (75.8%) | 2/14 (14.3%) |
| Q1–78 | 203/222 (91.4%) | 30/35 (85.7%) |
*Data for B lineage were not available before pass 56. 1
Three main products were consistently identified during the RAPD analysis. Table 2 shows that these three products were consistently found in all passes tested from the B and Q lineages, but not the J lineage. Although several other products were regularly seen, these three products were the most distinct and reproducible. No distinct, reproducible products were generated using the F15 primer. Figure 1 shows an example illustrating the distinction between these toxoplasma specific bands and other less reproducible bands in some of the pass numbers with primer B12.
Table 2.
Toxoplasm gondii specific random amplified polymorphic DNA product bands present in different lineage pass numbers
| Primer | Product size (bp) | Band presence (+) in lineage/pass number | ||||||||
| B21 | B79 | B192 | J4 | J21 | J79 | Q4 | Q22 | Q78 | ||
| B05 | 442 | + | + | + | + | + | − | + | + | + |
| B12 | 243 | + | + | + | − | − | + | + | + | + |
| B12 | 584 | + | + | + | − | + | + | + | + | + |
Figure 1.
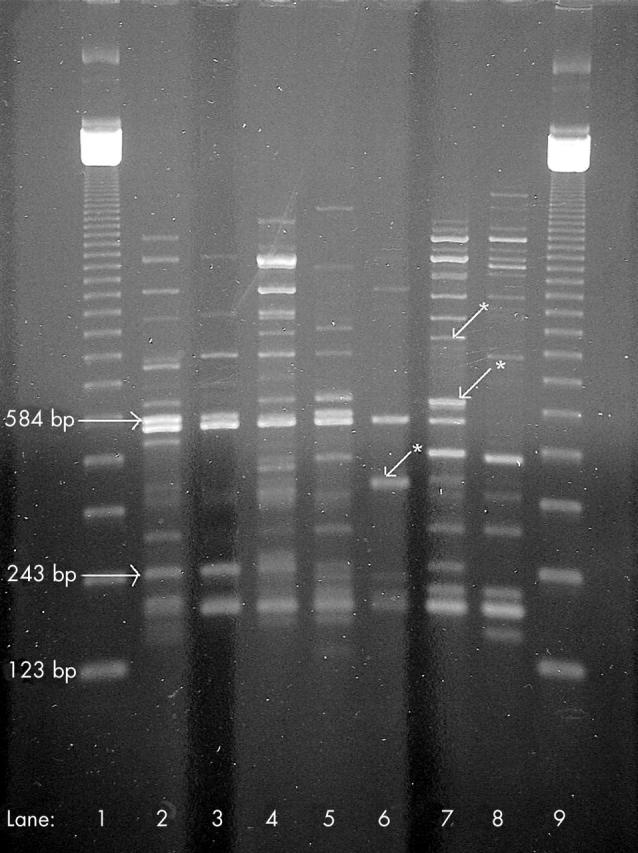
Random amplified polymorphic DNA analysis results using primer B12 on B, J, and Q lineage tachyzoites. Lanes 1 and 9, 123 bp marker; lanes 2 and 3, B lineage passes 21 and 79; lanes 4 and 5, Q lineage passes 22 and 78; lanes 6 and 7, J lineage passes 21 and 79; lane 8, uninfected cell control. Reproducible Toxoplasma gondii specific products (absent in uninfected cell control) of 243 bp and 584 bp are identified. *Other T gondii specific bands not found in at least four of the six RAPD runs, and were thus not included in the final analysis.
DISCUSSION
To be of use to the diagnostic laboratory a tachyzoite harvest success rate of > 90% is desirable. Initially, the Q lineage was the only one that regularly fulfilled these criteria. However, the B lineage was the first to be successfully incorporated into the continuous cell culture system, and overcome the practical problems in the system. 5 Early failures may therefore not have been a function of the lineage itself. Later passes (100 to 188) with the B lineage did fulfil the harvest success criteria on a significantly more regular basis (p < 0.001). Success in the dye test is crucial. B and Q lineage tachyzoites were successfully incorporated into the dye test, consistently producing good results. In contrast, those from the J lineage performed badly. We have considered only RAPD product bands that are distinct and consistently present, so our number of bands in the final analysis is small. However, two of these products (243 bp with the B12 primer and 442 bp with the B05 primer) are the same size as markers identified when Guo et al used the same RAPD method on DNA to distinguish between virulent and avirulent strains of T gondii. 8 Interestingly, using their typing method, the B, J, and Q lineages of the virulent RH strain appear to produce both virulent (243 bp product) and avirulent (442 bp product) markers, as long as it can be assumed that the same primers will produce the same bands using cDNA instead of DNA. This may suggest that gene expression has evolved with repeated passage in rats and continuous cell culture.
Take home messages .
Toxoplasma gondii B and Q lineages consistently produced tachyzoites that were successfully used in the dye test and their gene expression was stable after multiple passages
In contrast, the J lineage had unpredictable growth, tachyzoites unsuitable for use in the dye test, and changing gene expression with multiple passages
This study has explained some anomalies in the performance of different stocks of T gondii
The use of lineages that are still evolving should be avoided in diagnostic tests
“The irregular expression of the virulence and avirulence markers seen with the J lineage may indicate differential expression of the genes that are essential for adapting to cell culture”
The observation that the B and Q lineages appear to have adapted to growth in cell culture is substantiated by the expression of the three main gene products not altering with multiple passage. The irregular expression of the virulence and avirulence markers seen with the J lineage may indicate differential expression of the genes that are essential for adapting to cell culture, resulting in unpredictable growth and the production of tachyzoites unsuitable for use in diagnostic assays. The B, J, and Q lineages originated from stock that had previously been passaged in rats for many years. This material came from at least three different laboratories and was routinely passed through mice after approximately 50 passes to restore and maintain its virulence. The apparent initial lack of the virulence marker in the J lineage may be a function of the number of successive rat passes immediately before storage in liquid nitrogen. It may also simply be because it came from rat stock that had diverged at a different rate as a result of a fewer or greater number of passages, or had developed different polymorphisms. 3 Our study has explained some anomalies in the performance of different stocks of toxoplasma, and suggests that lineages that are still evolving in cell culture should be avoided.
Acknowledgments
We would like to thank the national services division of the Common Services Agency for the National Health Service in Scotland for the reference laboratory funding. S Mavin was a trainee clinical scientist funded by the National Services Division.
Abbreviations
PCR, polymerase chain reaction
RAPD, random amplified polymorphic DNA
TBE, Tris borate EDTA
REFERENCES
- 1. Ashburn D, Evans R, Chatterton, et al. Toxoplasma dye test using cell culture derived tachyzoites. J Clin Pathol 2000;53:630–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Evans R, Chatterton JMW, Ashburn D, et al. Cell-culture system for continuous production of Toxoplasma gondii tachyzoites. Eur J Clin Microbiol Infect Dis 1999;18:879–84. [DOI] [PubMed] [Google Scholar]
- 3. Howe DK, Sibley LD. Toxoplasma gondii: analysis of different laboratory stocks of the RH strain reveals genetic heterogeneity. Exp Parasitol 1994;78:242–5. [DOI] [PubMed] [Google Scholar]
- 4. Guo ZG, Johnson AM. Genetic characterization of Toxoplasma gondii strains by random amplified polymorphic DNA polymerase chain reaction. Parasitology 1995;111:127–32. [DOI] [PubMed] [Google Scholar]
- 5. Chatterton JMW, Evans R, Ashburn D, et al. Toxoplasma gondii in vitro culture for experimentation. J Microbiol Methods 2002;51:331–5. [DOI] [PubMed] [Google Scholar]
- 6. Jakobsen KS, Haugen M, Saebøe-Larssen S, et al. Direct mRNA isolation using magnetic oligo (dT) beads: a protocol for all types of cell cultures, animal and plant tissues. In: Ed Uhlén M, Hornes E, Olsvik, eds. Advances in biomagnetic separation. USA: Eaton Publishing, 1994:61–72.
- 7. Burg JL, Grover CM, Pouletty P, et al. Direct and sensitive detection of a pathogenic protozoan, Toxoplasma gondii by polymerase chain reaction. J Clin Microbiol 1989;27:1787–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo ZG, Gross U, Johnson AM. Toxoplasma gondii virulence markers identified by random amplified polymorphic DNA polymerase chain reaction. Parasitol Res 1997;83:458–63. [DOI] [PubMed] [Google Scholar]


