Abstract
Aims: To determine the nucleotide sequences of adenovirus (Ad) types 1 and 6 fibre genes; to clarify the molecular basis of the distinct haemagglutination properties of subgenus C Ads and their phylogenetic relations.
Methods: Human Ad1 and Ad6 fibre genes were sequenced from genomic DNA by direct sequencing. Primer selection was based on alignment of the fibre gene of human Ad serotypes Ad2 and Ad5. Fibre based subgenus C specific polymerase chain reaction (PCR) was performed to check for deletions in field isolates of Ad6, as revealed by sequence analysis of the Ad6 prototype. A phylogenetic tree was constructed from the predicted amino acid (AA) sequences of the fibre gene of important Ads.
Results: Ad1 and Ad6 comprise 1746 and 1584 nucleotides, encoding 582 and 528 AA, respectively. Ad6 showed deletions in motifs 15–17 (51 AA) of the shaft when compared with Ad1, Ad2, and Ad5. Subgenus C specific PCR with both prototype and field isolates also showed deletions in Ad6. In the shaft and knob, AA homology was 58.82–72.91% and 68.89–74.59%, respectively. The tail was 100% conserved. Phylogenetically, Ad1 and Ad6, including Ad2 and Ad5, formed a subgenus specific cluster, like other serotypes.
Conclusions: The fibre gene (including the knob region) of subgenus C Ads is heterogeneous, providing the molecular basis for lack of crossreactivity in the haemagglutination inhibition test. This heterogeneity could be helpful in fibre based genotyping of subgenus C field isolates. Phylogeny might be useful for subgenus specific identification of important field strains.
Keywords: subgenus C adenovirus, fibre gene, sequence, polymerase chain reaction, phylogenesis
In total, 51 adenovirus (Ad) serotypes have been identified as human pathogens, and on the basis of biochemical, immunological, and morphological criteria they are divided into six subgenera A–F. 1, 2 The members of subgenus C—Ad1, Ad2, Ad5, and Ad6—are the most frequently isolated human Ads, causing respiratory infection, gastroenteritis with prolonged excretion in children, ocular disease, and opportunistic infection among immunocompromised patients. 3– 5 Restriction endonuclease analysis of subgenus C field isolates revealed bewildering variations in their genome. 6– 9 Surprisingly, they have distinct antigenic sites for serum neutralisation (SN) and haemagglutination inhibition (HAI). 10, 11 The fibre protein, one of the major capsid proteins, is responsible for haemagglutination and the attachment of Ads to a specific cellular receptor at the early stage of infection through a receptor binding site present in the knob region. 12, 13 Analysis of the fibre protein of human Ad2 revealed that it possesses three distinct regions: an N-terminal tail of 43 amino acid (AA) residues; a central shaft of 357 residues, consisting of 22 pseudorepeats, usually 15 residues each (motif); and a C-terminal domain of 181 residues forming the knob. 14 A later study by Chrobzek and Jarcot (1987) 15 indicated that the tail comprises 44 AA residues. The tail and knob portions of the fibre are not significantly variable in length among the serotypes. However, the number of pseudorepeats in the shaft, which determines the length of the fibre, is variable. The tropism of Ads primarily depends on the presence of specific AA in the knob portion of the fibre. 13 Investigation have shown that, as the length of fibre shaft increases, the efficiency of viral attachment to the cellular receptor also increases. 16, 17 Although subgenus C Ads account for 59% of all infections caused by Ads, Ad6 is responsible for only 4% of these infections, compared with 34.3% for Ad1, 42.8% for Ad2, and 18.6% for Ad5. 4 Despite the fact that these subgenus C Ads are important human pathogens, the fibre genes of Ad1 and Ad6 are yet to be studied.
“The fibre protein is responsible for haemagglutination and the attachment of adenoviruses to a specific cellular receptor at the early stage of infection through a receptor binding site present in the knob region”
In our study, the full length fibre genes of both Ad1 and Ad6 prototypes were sequenced for the first time with the following aims: (a) to find possible variations in the Ad1 and Ad6 fibre genes compared with the other serotypes of subgenus C, and to confirm any variations by fibre based polymerase chain reaction (PCR); (b) to correlate the variations of subgenus C Ads in the HAI test with heterogeneity of the fibre protein in the knob region; and (c) to clarify any evolutionary relations between Ad1 and Ad6 using a phylogenetic method.
MATERIALS AND METHODS
Viruses and DNA
Ad prototype strains Ad1 (adenoid 71), Ad2 (adenoid 6), Ad5 (adenoid 75), and Ad6 (tonsil 99) and serotypes of other subgenus Ads were obtained from the American Type Culture Collection (ATCC; Rockville, Maryland, USA). Seventeen geographically and temporally diverse field isolates (Ad1:4, Ad2:4, Ad5:4, and Ad6:5) previously typed by the SN were used in our study.
Extraction of viral DNA
The viruses were passaged in A549 cells and DNA was extracted from the infected cells by the method described previously. 18 In short, when a greater than 75% cytopathic effect was seen, the cells were dislodged and pelleted by low speed centrifugation, washed three times with phosphate buffered saline, and resuspended in lysis buffer (10mM Tris/HCl, pH 7.4, 10mM EDTA, 1% sodium dodecyl sulfate). Next, the suspension was incubated with proteinase K (Sigma Chemical, St Louis, Missouri, USA). Cellular DNA was precipitated with 5M NaCl, and pelleted by centrifugation. After incubation with RNase A (Sigma Chemical) the supernatant was extracted twice in phenol/chloroform and precipitated in two volumes of absolute ethanol. After drying, DNA was suspended in 50 μl of double distilled water.
DNA sequencing
The genomic DNA was used as a template for DNA sequencing. The initial selection of primers was based on alignment of the Ad2 and Ad5 fibre genes (GenBank accession numbers j01907 (Ad2) and m18369 (Ad5)), and subsequently, because of the heterogeneous nucleotide sequences, the primer walking method was used. 15 DNA sequencing was performed on the sense and antisense strands with overlapping primers. The cycle sequence reaction was carried out with an ABI prism big dye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, California, USA). The sequences were read by Genetic analyser model 310 (Applied Biosystems). The nucleotide sequences of the fibre gene were translated into AA sequences, and compared with the published AA sequences of Ad2 and Ad5 using the GENETYX-MAC program (Software Development Co, Tokyo, Japan).
Nucleotide sequence accession numbers
The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank nucleotide sequence database with the accession numbers ab125750 (Ad1) and ab125751 (Ad6).
Ad subgenus C specific primers
Subgenus C specific PCR was performed with prototypes and field isolates to check the deletion in the shaft region of Ad6 field isolates noted by sequence analysis of the Ad6 prototypes. The design of the subgenus C specific primers (AdnCF, 5′-TGC TTG CGC THA AAA TGG GC-3′ (654–674) and AdnCR, 5′-CGA TTC TTT ATT CYT GGG CAA TGT-3′ (2228–2205)) was based on the alignment of the fibre gene sequences of Ad1 and Ad6 conducted in our laboratory (GenBank accession numbers Ad1 (ab125750), and Ad6 (ab125751)) and Ad2 and Ad5 obtained from GenBank (accession numbers Ad2 (j01917) and Ad5 (m18369)). 15 The nucleotide positions of the primers refer to the fibre sequence of Ad2.
DNA amplification and detection
PCR amplification was carried out in a 50 μl reaction mixture containing 1 μl aliquots of DNA, 5 μl of 10× concentrated buffer, 0.5 μM of each of the primers, 200 μM of each deoxynucleoside triphosphate (dNTP), and 1.25 U of Taq polymerase (Boehringer-Mannheim, Mannheim, Germany). Sterile distilled water was used as a negative control. For field isolates, Ad2 DNA was used as a positive control. The assay was performed in a thermal cycler (model 9600-R; Perkin Elmer, Foster City, California, USA). Thermal cycling consisted of preliminary denaturation for three minutes at 94°C, followed by 35 cycles of denaturation at 94°C for one minute, annealing at 47°C for one minute, and extension at 72°C for two minutes, with a final extension at 72°C for seven minutes. The reaction products (5 μl aliquots) were electrophoresed on a 1.2% horizontal agarose gel. After staining with ethidium bromide the gel was photographed with a CCD camera. The specificity of the test was evaluated with pooled DNA from the A, B, D, E, and F subgenera of Ad and non-adenoviral DNA.
Phylogenetic analysis
A phylogenetic tree was constructed according to the full length predicted AA sequences of the fibre genes obtained from the GenBank. Sequence alignment was performed with Clustal X (http://www-igbmc.u-strasbg.fr/BioInfo/) with parameters provided in Clustal X. Evolutionary distances were estimated by Kimura’s two parameter method. A phylogenetic tree with 1000 bootstrap replicates was generated by the neighbour joining method with Clustal W and plotted by Tree View (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
RESULTS
Sequence analysis
The Ad1 and Ad6 fibre genes were 1746 and 1584 nucleotides (nt) long (table 1 ), corresponding to 582 and 528 AA, respectively. The predicted AA sequences were consistent with the structural domains described for Ad2, which possesses three distinct regions: an N-terminal tail (44 AA); a central shaft of 357 residues consisting of 22 pseudorepeats, usually 15 residues; and a C-terminal knob of 181 residues. The fibre tail of Ad1 and Ad6 was 132 nt long, corresponding to 44 AA, and showed 100% conservation among the members of subgenus C. However, at the nucleotide level the homology varied from 89.39% to 93.18%. The two conserved sequences—2-KRλR (where λ is A in all the subgenus C serotypes) and FNPVYPYD—are present in subgenus C, similar to the other Ad subgenera (fig 1 ). 19
Table 1.
Length of the fibre genes (number of nucleotides) and proteins (number of amino acids) among the members of subgenus C adenoviruses
| Tail | Shaft | Knob | Overall | ||
| Ad1 | DNA | 132 | 1071 | 543 | 1746 |
| Protein | 44 | 357 | 181 | 582 | |
| Ad2 | DNA | 132 | 1071 | 543 | 1746 |
| Protein | 44 | 357 | 181 | 582 | |
| Ad5 | DNA | 132 | 1071 | 540 | 1743 |
| Protein | 44 | 357 | 180 | 581 | |
| Ad6 | DNA | 132 | 918 | 534 | 1584 |
| Protein | 44 | 306 | 178 | 528 |
Ad, adenovirus.
Figure 1.

Comparison of predicted amino acid (AA) sequences of adenovirus 1 (Ad1), Ad2, Ad5, and Ad6 fibre proteins. The nucleotide sequences of Ad1 and Ad6 have been deduced from their respective DNA sequences. Ad2 and Ad5 AA sequences were obtained from GenBank using the accession numbers j01917 and m18369, respectively. The conserved residues in the tail are underlined. The four sequences have been aligned to obtain maximal homology. The structural domains tail, shaft, and knob are marked. The shaft region has been divided into motifs. Bold letters indicate hydrophobic residues.
The shaft region of Ad1 and Ad6 displayed a 15 AA basic structural unit (motif), containing hydrophobic glycine (G) or proline (P) residues at position 8 of the β sheet/β turn model (fig 1 ). 14 Ad1 comprises 357 polypeptides and displayed 22 repeats of the 15 AA motifs, similar to Ad2 and Ad5. The shaft of Ad6 was relatively shorter, with 306 AA (19 motifs) as a result of the deletion of motifs 15 to 17 (fig 2 ). The third motifs were 19 AA long except for in Ad6, which showed deletion of a residue (T) in position 8. In the shaft region, AA homology among the serotypes varied from 58.82% to 72.91%. The Ad1 and Ad6 fibre knobs were 543 and 534 nt long, corresponding to 181 and 178 AA, respectively. Their AA homology varied between 68.89% and 74.59% (table 2 ). Ad1 and Ad6 showed an overall homology of 71.95% at the DNA level and 70.1% at the AA level. The overall AA homology among the members of subgenus C varied from 65.06% to 75.47%, and is considered to be heterogeneous.
Figure 2.
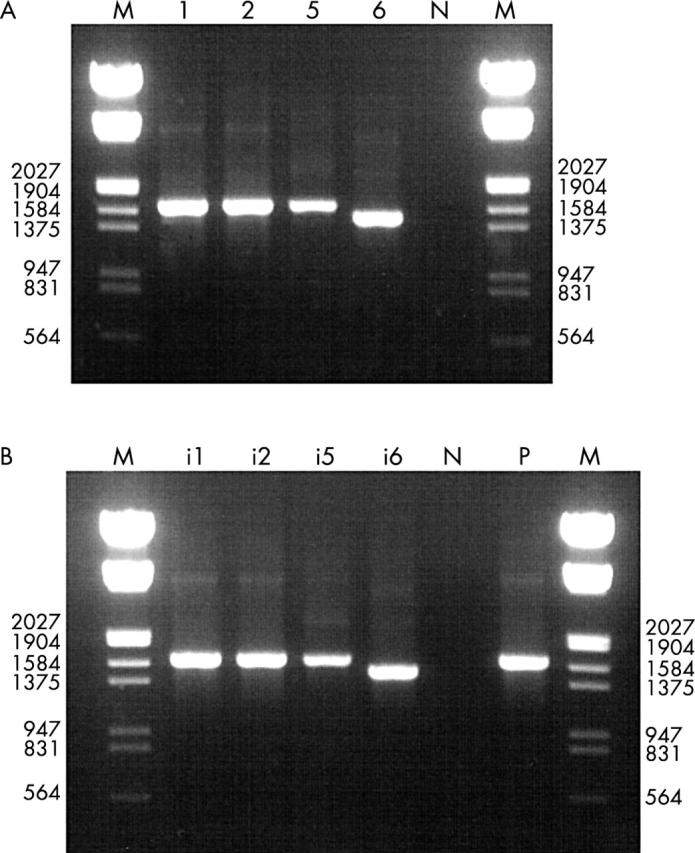
(A) Ethidium bromide stained agarose gel showing polymerase chain reaction (PCR) products of subgenus C prototype strains. The lane designations indicate the respective prototypes: lane N, negative control; lane M, DNA molecular size marker III (Boehringer Manheim). (B) Ethidium bromide stained agarose gel showing PCR products of subgenus C field isolates. Lane i1 (66–315), Ad1; lane i2 (YC 66–283), Ad2; lane i5 (YC 77–82), Ad5; lane i6 (YC 92-516T), Ad6; lane N, negative control; lane P, positive control (Ad2p); lane M, DNA molecular size marker III.
Table 2.
Fibre gene nucleotide and amino acid homologies (%) among the members of the subgenus C adenoviruses
| Fibre | Ad1/2 | Ad1/5 | Ad1/6 | Ad6/2 | Ad6/5 | |
| Tail | DNA | 90.15 | 90.91 | 93.18 | 90.91 | 89.39 |
| Protein | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | |
| Shaft | DNA | 72.71 | 74.91 | 69.16 | 64.52 | 63.90 |
| Protein | 70.56 | 72.91 | 64.71 | 62.18 | 58.82 | |
| Knob | DNA | 68.56 | 72.56 | 72.24 | 73.85 | 69.98 |
| Protein | 70.88 | 74.59 | 73.48 | 74.03 | 68.89 | |
| Overall | DNA | 72.73 | 75.39 | 71.95 | 69.42 | 67.72 |
| Protein | 72.87 | 75.47 | 70.10 | 68.73 | 65.06 |
DNA and amino acid homologies were obtained using GENETYX-MAC software (Software Development Co, Tokyo, Japan).
Ad, adenovirus.
Polymerase chain reaction
The primer pair AdnCF/AdnCR amplified approximately 1575 bp, 1575 bp, and 1572 bp products from Ad1, Ad2, Ad5 prototypes, respectively, whereas the Ad6 prototype yielded a 1413 bp product (fig 2A ). Seventeen field isolates (table 3 ) also yielded clearly visible products of the expected length (fig 2B ). Ad6 could be distinguished from the other members of subgenus C by the length of the amplicon (1413 bp). PCR using DNA from other subgenera of adenovirus and non-adenoviral DNA yielded no amplified products, indicating a high specificity of the test.
Table 3.
Fibre based subgenus C specific polymerase chain reaction
| Original identification | Subgenus/amplicon (bp) | |
| Isolate | Serotype | |
| YC 66-113 | Ad1 | C/1572-1575 |
| YC 66-283 | Ad2 | C/1572-1575 |
| YC 66-284 | Ad2 | C/1572-1575 |
| YC 66-315 | Ad1 | C/1572-1575 |
| YC 72-141 | Ad5 | C/1572-1575 |
| YC 72-143 | Ad2 | C/1572-1575 |
| YC 77-82 | Ad5 | C/1572-1575 |
| YC 84-31 | Ad6 | C/1413 |
| YC 84-37 | Ad5 | C/1572-1575 |
| YC 85-70 | Ad6 | C/1413 |
| YC 85-75 | Ad1 | C/1572-1575 |
| YC 85-77 | Ad1 | C/1572-1575 |
| 92-515T | Ad5 | C/1572-1575 |
| 91-1141 | Ad6 | C/1413 |
| 92-516T | Ad6 | C/1413 |
| 92-523T | Ad6 | C/1413 |
| 92-566T | Ad2 | C/1572-1575 |
Ad, adenovirus.
Phylogenetic analysis
Ad1 and Ad6 were included in a cluster formed by subgenus C adenoviruses. Ad6 was found to be more closely related to Ad1 than either Ad2 or Ad5. Other serotypes included in phylogeny also formed subgenus specific clusters. However, the cluster formed by subgenus F was divided into two subclusters because of the presence of two different fibres (long fibre and short fibres; fig 3 ).
Figure 3.
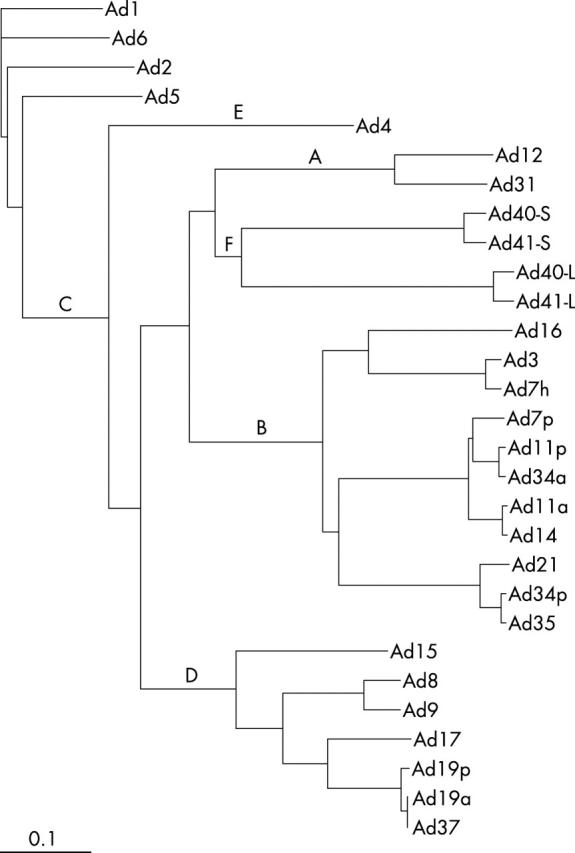
A phylogram showing adenovirus (Ad) subgenus specific clusters indicated by letters A–F. Ad1 and Ad6 are included in cluster C. The subgenus F is divided into short and long fibre subclusters marked by L and S with the respective prototypes. The phylogenetic tree was reconstructed from the predicted amino acid sequences of the full length fibre gene of all important human adenovirus serotypes under the GenBank accession for subgenus A: Ad12 (x73487) and Ad31 (x76548); subgenus B: Ad3 (m12411), Ad7p (m23696), Ad7h (z48954), Ad11p (l08231), Ad11a (l08232), Ad14 (ab065116), Ad16 (u06106), Ad21 (u06107), Ad34p (u10271, ab073168), Ad34a (u10272), and Ad35 (u10272); subgenus C: Ad1 (ab125750), Ad2 (j01917), Ad5 (m18369), and Ad6 (ab125751); subgenus D: Ad8 (x74660), Ad9 (x74659), Ad15 (x72934), Ad17 (nc002067), Ad19p (u69130), Ad19a (u69131), and Ad37 (u69132); subgenus E: Ad4 (x76547); and subgenus F: Ad40 (l19443 and m28822) and Ad41 (x16583, m60327).
DISCUSSION
Subgenus C Ads differ from other subgenera in that they have a highly variable genomic organisation, they are in widespread use as a vector for gene delivery, and they are responsible for approximately 60% of human infections caused by the different Ad serotypes. Our study revealed two important findings about the subgenus C Ad fibre protein. First, the structure of the knob is heterogeneous, which could explain the serological characteristics of subgenus C Ads and, second, the fibre shaft of Ad6 is relatively short, and this may be related to its lower rate of infection.
Subgenus C Ads are unique because of the lack of crossreactivity in both the SN and HAI tests. 20 The HAI test in a large number of geographically diverse isolates showed that the fibre epitopes are relatively conserved, although the exact location and size of these epitopes are yet to be determined. 10, 11, 21 Serotypes such as Ad40 and Ad41 or Ad19 and Ad37 show 97.7% and 95.5% homology in the knob region, respectively, and show major crossreactivity in the HAI test. Therefore, higher homology (> 80%) in the knob region is thought to be the basis of serological crossreactivity as a result of epitope sharing. 19, 20, 22 Although not an absolute rule, lower homology in the knob region may imply the presence of distinct epitopes, as demonstrated by the absence of major crossreactivity in HAI. Thus, subgenus C Ads have no major crossreactvity in HAI with other subgenera or among themselves probably because they have low homology in the fibre knob region, between 68.89% and 74.59.
“Adenovirus 1 (Ad1) is more closely related to Ad6 than to Ad2 or Ad5”
To date, studies with Ad2 and Ad5 have shown that the subgenus C fibre knob mediates binding to the coxsackie virus and Ad receptor (CAR), and is thus a major determinant of viral tropism. 23 Mounting evidence suggests that at least two factors determine viral infectivity and tropism, namely: (a) the fibre knob–primary receptor interaction and (b) the length and flexibility of the fibre shaft. 16, 17 The length of the fibre shaft is variable because of deletion or insertion of certain numbers of repeats. The formation of a recombinant fibre shaft with decreasing length and increasing rigidity significantly reduced infectivity and attachment of Ad5. 24 The major differences between the Ad6 fibre shaft and that of the other subgenus C Ads are the deletion of the 15–17th motifs (51 residues) of the shaft and a single residue (T) deletion in the eighth position of motif 3. The deletion was also detected in PCR products of the field isolates of Ad6. It may be possible that deletion of 51 AA residues in the shaft decreases its flexibility and reduces the mobility of the knob. 22 The third motif of subgenus C is 19 AA long except in Ad6. In all Ads, the third motif carries a hydrophilic residue in the second hydrophobic position of the β strand (position 15 in subgenus C). The fibre bends above the penton base at a position corresponding to this third motif of the shaft, which is also a flexing point to allow fibre movement. In Ad6, the third motif is 18 AA long (rather than 19 AA as in other subgenus C Ads), as a result of the deletion of a single residue (T) at position 8, and this may also slightly decrease the flexibility of the fibre shaft. 25 Both these factors present in Ad6 might act together to decrease its efficiency of binding to the cellular receptor. Further experiments with recombinant Ad6 fibre proteins might be useful to investigate the relation between the length of the Ad6 fibre shaft and the rate of infection. It is also important to investigate whether Ad6 uses CAR or another cellular receptor.
Take home messages .
The fibre gene (including the knob region) of subgenus C adenoviruses (Ad) is heterogeneous, providing the molecular basis for their lack of crossreactivity in the haemagglutination inhibition test
This heterogeneity might be useful in fibre based genotyping of subgenus C field isolates
Phylogeny might be useful for subgenus specific identification of important field strains
The fibre shaft of Ad6 is relatively short, and this may be related to its lower rate of infection
In the phylogram, Ad1 and Ad6, together with Ad2 and Ad5, formed a subgenus C specific cluster, indicating that these Ads have the same ancestral origin. Ad1 is more closely related to Ad6 than to Ad2 or Ad5. Subgenus specific clusters were also formed by other serotypes as a result of high homology (60–80%) among the members of each subgenus. 26 Subgenus specific clusters of the fibre gene that included important Ads could be useful in subgrouping and understanding the evolution of Ads.
Sequence heterogeneity in the fibre knob provides the molecular basis of the distinct serological properties of subgenus C Ads in the HAI test. The inclusion of serotypes Ad1, Ad2, Ad5, and Ad6 in a single cluster defines the evolutionary relations between the subgenus C Ads. Furthermore, the sequence data of Ad1 and Ad6 reported in our study will be useful in the identification of subgenus C field strains, either by fibre based type specific PCR or by comparison of sequences.
Acknowledgments
Supported by grant-in-aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS), Japan.
Abbreviations
AA, amino acids
Ad, adenovirus
CAR, coxsackie virus and adenovirus receptor
HAI, haemagglutination inhibition
nt, nucleotides
PCR, polymerase chain reaction
SN, serum neutralisation
REFERENCES
- 1. Shenk T. Adenoviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, eds. Fields virology. 5th ed. Philadelphia: Lippincott-Raven Publishers, 1996:2111–48.
- 2. De Jong JC, Wermenbol AG, Verweiji-Uijterwall, et al. Adenovirus from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotype Ad50 and Ad51 of species B1 and D, respectively. J Clin Microbiol 1999;37:3940–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fox JP, Hall CE, Cooney MK. The Seattle virus watch. VII. Observation of adenovirus infection. Am J Epidemiol 1977;105:362–68. [DOI] [PubMed] [Google Scholar]
- 4. Schmitz H, Wigand R, Heinreich W. World wide epidemiology of adenovirus infections. Am J Epidemiol 1983;117:455–66. [DOI] [PubMed] [Google Scholar]
- 5. Hierholzer JC. Adenoviruses in the immunocompromised host. J Clin Microbiol 1992;5:262–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adrian T. Genome polymorphism of human adenovirus subgenus C. Arch Virol 1996;141:1021–31. [DOI] [PubMed] [Google Scholar]
- 7. Adrian T, Wigand R, Wadell G. Serological and biochemical characteristics of intermediate adenovirus strains of subgenus D. Arch Virol 1987;97:347–57. [DOI] [PubMed] [Google Scholar]
- 8. Adrian T, Wolf U. Adenovirus 6 genome types: mapping of restriction site alterations on the genome. Res Virol 1989;140:545–50. [DOI] [PubMed] [Google Scholar]
- 9. Fife KH, Ashley R, Corey L. Isolation and characterization of six new genome types of human adenovirus types 1 and 2. J Clin Microbiol 1985;21:20–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Adam E, Nasz I, Lengyel A. Antigenic homogeneity among the members of the adenovirus hexon types of subgenus C. Arch Virol 1995;140:1297–301. [DOI] [PubMed] [Google Scholar]
- 11. Kaier B, Wigand R. Antigenic homogeneity of adenovirus type 1, 2, 5, and 6. J Med Virol 1986;18:283–7. [DOI] [PubMed] [Google Scholar]
- 12. Norby E. The structural and functional diversity of adenovirus capsid components. J Gen Virol 1969;5:221–36. [DOI] [PubMed] [Google Scholar]
- 13. Arnberg N, Mei Y-F, Wadell G. Fiber genes of adenoviruses with tropism for the eye and the genital tract. Virology 1997;227:239–44. [DOI] [PubMed] [Google Scholar]
- 14. Green NM, Wrigley NG, Russell WC. Evidence for a repeating cross-β sheet structure in the adenovirus fiber. EMBO J 1983;2:1357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chrobozek J, Jacrot B. The sequence of adenovirus fiber: similarities and differences between serotypes 2 and 5. Virology 1987;161:549–54. [DOI] [PubMed] [Google Scholar]
- 16. Shayakhmetov DM, Lieber A. Dependence of adenovirus infectivity on length of the fiber shaft domain. J Virol 2003;74:10274–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nemerow GR. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J Virol 2000;77:7225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adhikary AK, Numaga J, Kaburaki T, et al. Genetic characterization of adenovirus type 8 isolated in Hiroshima city over a 15 year period. J Clin Pathol 2003;56:120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pring-Akerblom P, Heim A, Trijssenaar FEJ. Conserved sequences in the fibers of epidemic keratoconjunctivitis associated human adenovirus. Arch Virol 1997;142:205–11. [DOI] [PubMed] [Google Scholar]
- 20. Hierholzer JC. Antigenic relationship among the 47 human adenoviruses determined in reference horse antisera. Arch Virol 1991;121:179–97. [DOI] [PubMed] [Google Scholar]
- 21. Adhikary AK, Inada T, Suzuki E, et al. Characterization of hexon and fibre gene of a novel strain of adenovirus involved in epidemic keratoconjunctivitis. J Clin Pathol 2004;57:95–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kidd AH, Erasmus MJ, Tiemessen CT. Fiber sequence heterogeneity in subgroup F adenoviruses. Virology 1990;179:139–50. [DOI] [PubMed] [Google Scholar]
- 23. Bergelson DM, Cunningham JA, Droguett G, et al. Isolation of common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 1997;275:1320–3. [DOI] [PubMed] [Google Scholar]
- 24. Nakamura T, Sato K, Hamada H. Reduction of natural adenovirus tropism to the liver by both ablation of fiber-coxsackie virus and adenovirus receptor interaction and use of replaceable short fiber. J Virol 2003;77:2512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ruigrok RWH, Barge A, Mittal SK, et al. The fiber of bovine adenovirus type 3 is very long but bent. J Gen Virol 1994;75:2069–73. [DOI] [PubMed] [Google Scholar]
- 26. Pring-Akerblom P, Adrian T. Sequence characterization of the adenovirus 31 fiber and comparison with serotypes of subgenera A to F. Res Virol 1995;146:343–54. [DOI] [PubMed] [Google Scholar]


