Abstract
Background: The isolation of good quality DNA from routinely fixed and processed biopsy samples is crucial for the success of subsequent molecular analysis.
Aims: To compare the amount of β actin DNA extracted from upper gastrointestinal tract biopsies fixed in buffered and unbuffered formalin.
Methods: Amounts of β actin DNA extracted from forceps biopsies of the upper gastrointestinal tract fixed in unbuffered (n = 22) and buffered formalin (n = 16) were estimated by quantitative real-time polymerase chain reaction.
Results: The yield of β actin DNA was significantly higher in biopsies fixed in buffered formalin than in those fixed in unbuffered formalin (median 2.8 × 104 and 5.3 × 102 DNA molecules, respectively; p < 0.005). Furthermore, fixation in buffered formalin led to a more reproducible DNA extraction, as indicated by the coefficient of variation (1.0 and 2.2, respectively).
Conclusions: This study indicates that tissue samples should be fixed in buffered formalin to facilitate the use of molecular pathology analysis in routine biopsy material.
Keywords: unbuffered formalin fixation, gastric and duodenal biopsy, paraffin wax embedding, real-time (TaqMan) polymerase chain reaction, human β actin
The isolation of DNA from routinely formalin fixed and paraffin wax embedded tissue samples permits the use of archival materials for molecular analysis. A major problem with these specimens is the degradation of nucleic acids, which could lead to false negative results.
In formalin fixed tissue several factors affect the degradation of DNA, including the duration of fixation, pH, salt concentration, and temperature. 1– 8
“The isolation of DNA from routinely formalin fixed and paraffin wax embedded tissue samples permits the use of archival materials for molecular analysis”
In our present study, we compared the effect of buffered and unbuffered formalin fixation on routinely processed small forceps biopsies of the upper gastrointestinal tract using real-time polymerase chain reaction (PCR) to measure the commonly used control gene human β actin. We further compared DNA amplification of gastric and duodenal biopsies to explore the possibility that gastric acid might increase DNA degradation.
MATERIALS AND METHODS
Routinely processed gastric and duodenal biopsy samples were chosen from the paraffin wax embedded archived material of the Cantonal Institute of Pathology in Liestal, Switzerland. Because the institute receives material from various institutions using different types of formalin fixatives, the tissue blocks were selected depending on the use of buffered or unbuffered formalin. All tissue samples were immediately immersed in the fixation solution and the total duration of fixation was less than two days.
Fifty 2 μm thick sections were collected from each paraffin wax block in a microtube. DNA was extracted with the QIAmp DNA mini kit (Qiagen AG, Basel, Switzerland) according to the manufacturer’s instructions. DNA was measured by spectrophotometry. A DNA fragment of the human β actin gene was amplified using TaqMan β actin control reagents according to the manufacturer’s instructions (catalogue number 401846; Applied Biosystems, Rotkreuz, Switzerland). The PCR reaction mix contained 100–500 ng of sample DNA, 25 μl Universal master mix (catalogue number 4304437; Applied Biosystems), 5 μl of each primer (final concentration 3μM), and 5 μl β actin probe (final concentration 200nM) in a total volume of 50 μl. Samples were placed in the MicroAmp 96 well reaction plate and covered with MicroAmp optical caps (catalogue number N-801-0935; PE Biosystems, Rotkreuz, Switzerland). TaqMan PCR was performed on the ABI Prism 7700 sequence detection system (Applied Biosystems) under the following conditions: activation of the Ampli Taq Gold DNA polymerase at 95°C for 10 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds, primer annealing and extension at 60°C for 60 seconds. The size of the PCR product is 295 bp. All biopsies were tested in duplicate.
The β actin PCR product was cloned into the PCR-Script Amp SK(+) plasmid (Invitrogen, Basel, Switzerland); plasmid DNA was measured by spectrophotometry and a serial dilution of 6 pg/μl (equivalent to 9 × 106 plasmid DNA molecules) to 0.006 pg/μl (9 × 103 plasmid DNA molecules) was tested. Herring sperm DNA was added if appropriate to keep a total (background) amount of 250 ng DNA/assay. Each analysis was done in triplicate.
Statistical analysis was performed using the Student’s unpaired t test and a p value of < 0.05 was considered significant.
RESULTS
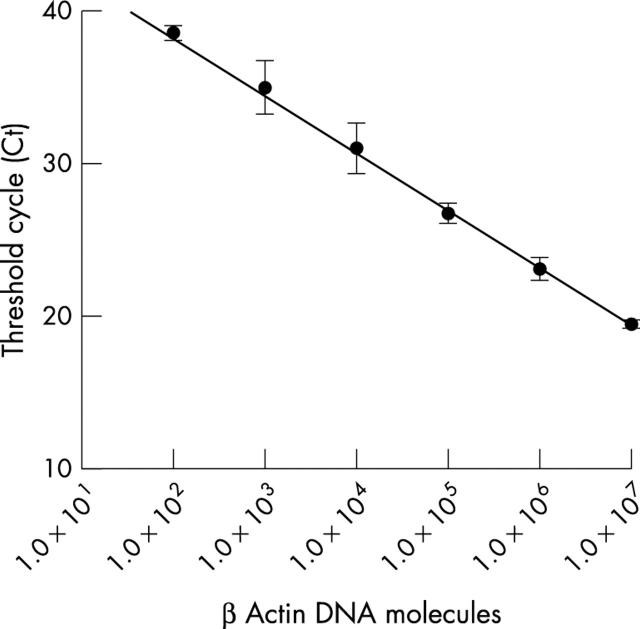
Using serial dilutions of human β actin plasmid DNA in 250 ng of herring sperm DNA, a wide linear association was seen and a sensitivity of about 40 β actin DNA molecules (equivalent to 20 cells) was reached (fig 1 ).
Figure 1.
Real-time polymerase chain reaction analysis of logarithmic serial dilutions from a plasmid DNA with a human β actin insert. The analysis was performed in triplicate and the standard deviations are indicated. The sensitivity of the assay is about 40 human β actin DNA molecules, equivalent to 20 cells.
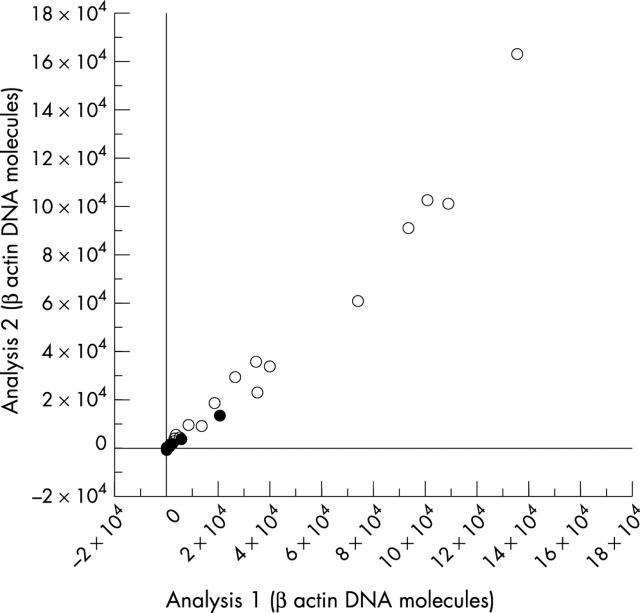
In total, 38 biopsy samples were analysed, including 19 gastric and 19 duodenal forceps biopsies. Twenty two biopsies had been fixed in buffered and 16 in unbuffered formalin. The mean β actin values of the 19 gastric and 19 duodenal biopsies were 1.7 × 104 DNA molecules (range, 5 to 1 × 105) and 2.1 × 104 (range, 15 to 1.5 × 105) DNA molecules, respectively (difference not significant). In contrast, the mean number of β actin DNA molecules extracted from biopsies fixed in buffered formalin was 4.4 × 104, which is significantly higher than the mean of 1.6 × 103 β actin DNA molecules obtained from the biopsies fixed in unbuffered formalin (p < 0.005). In addition, a much wider range of 5 to 1.6 × 104 β actin DNA molecules was seen for those biopsies fixed with unbuffered formalin than was seen for biopsies fixed in buffered formalin (2.8 × 103 to 1.5 × 105 β actin DNA molecules). This is further highlighted by the fact that the median and mean numbers of β actin DNA molecules were similar in the biopsies fixed in buffered formalin (median, 2.8 × 104), whereas in unbuffered formalin fixed tissue the median (5.3 × 102) was considerably lower than the mean (fig 2 ). A good correlation was seen in the duplicate analysis of single clinical samples with a wide range of DNA content (r s = 0.991; p < 0.0001; fig 3 ).
Figure 2.
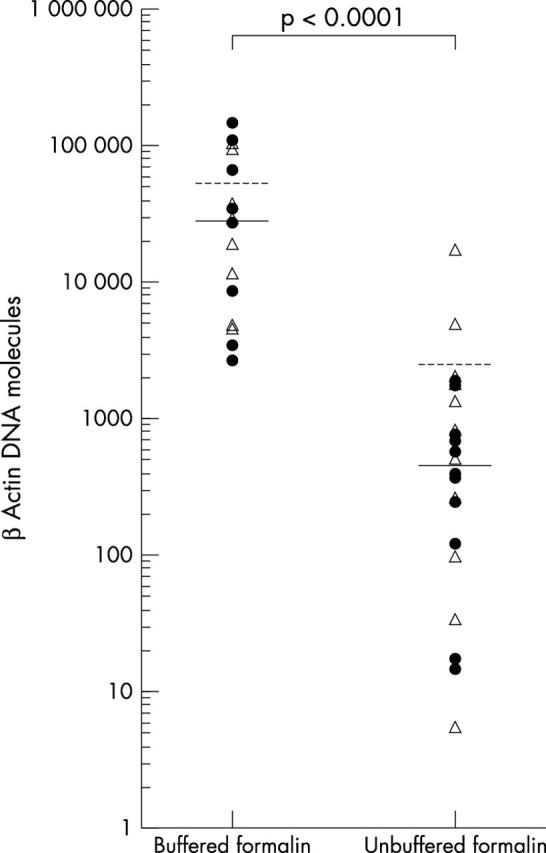
Real time polymerase chain reaction of human β actin DNA molecules extracted from biopsies fixed in buffered and unbuffered formalin. The difference between the two groups is significant (Student’s t test p < 0.0001). The median values are indicated by the solid lines and the mean values by the broken lines. The open triangles represent duodenal biopsies and the closed circles gastric biopsies.
Figure 3.
Correlation of the amount of human β actin DNA molecules in duplicate analyses of clinical samples. The numbers of DNA molecules are given in 104 β actin DNA molecules. Open circles represent samples fixed in buffered formalin and closed circles represent samples fixed in unbuffered formalin.
Using the same conditions, similar results were obtained after storing the DNA for four weeks at 4°C (data not shown).
DISCUSSION
Our study demonstrated the favourable impact of buffered formalin on the preservation of DNA in routinely embedded tissue samples. Using real-time PCR, we showed that up to 50 times more β actin DNA molecules could be extracted from small forceps biopsy samples fixed in buffered formalin compared with those fixed in unbuffered formalin. Furthermore, fixation in buffered formalin led to a more reproducible DNA extraction, as indicated by a narrower range of DNA molecules for each biopsy and a lower coefficient of variation (fig 2 ).
“Up to 50 times more β actin DNA molecules could be extracted from small forceps biopsy samples fixed in buffered formalin compared with those fixed in unbuffered formalin”
Formalin is the most widely used fixative in histopathological practice because it is easy to handle, shows low toxicity, is inexpensive, leads to good quality of morphology, and a large number of antibodies can be applied for immunohistochemistry. For molecular analysis, the duration of fixation is the major factor affecting the preservation of DNA in formalin fixed tissue. 1, 2, 4– 7 Unbuffered formalin oxidises to formic acid and an acidic environment causes degradation of nucleic acids because the β glycosidic bonds in the purine bases are hydrolysed at pH 4. 8 However, as shown by our gastric biopsies, low pH in the tissue itself does not have a negative effect on the quality of DNA. Furthermore, our data show that spectrophotometry does not reflect the quality of DNA extracted from paraffin wax embedded tissue.
Take home messages .
More β actin DNA could be extracted from biopsies fixed in buffered formalin than those fixed in unbuffered formalin and extraction was more reproducible in buffered formalin fixed tissue
Thus, tissue samples should be fixed in buffered formalin to facilitate the use of molecular pathology analysis in routine biopsy material
Although the buffered formalin is slightly more expensive, our study clearly demonstrates that tissue samples should be fixed in buffered formalin to facilitate the use of molecular analysis in routine biopsy material.
Abbreviations
PCR, polymerase chain reaction
REFERENCES
- 1. Karlsen F, Kalantari M, Chotemerere M, et al. Modifications of human and viral deoxyribonucleic acid by formaldehyde fixation. Lab Invest 1994;71:604–11. [PubMed] [Google Scholar]
- 2. Legrand B, Mazancourt P, Durigon M, et al. DNA genotyping of unbuffered formalin fixed paraffin embedded tissues. Forensic Sci Int 2002;125:205–11. [DOI] [PubMed] [Google Scholar]
- 3. Lie ES, Heyden A, Johannesen MKI, et al. Detection of human papillomavirus in routinely processed biopsy specimens from laryngeal papillomas: evaluation of reproducibility of polymerase chain reaction and DNA in situ hybridisation procedures. Acta Otolaryngol 1996;116:627–32. [DOI] [PubMed] [Google Scholar]
- 4. Alaibac M, Filotico R, Giannella C, et al. The effect of fixation type on DNA extracted from paraffin-embedded tissue for PCR studies in dermatopathology. Dermatology 1997;195:105–7. [DOI] [PubMed] [Google Scholar]
- 5. Ben-Ezra J, Johnson DA, Rossi J, et al. Effect of fixation on the amplification of nucleic acids from paraffin-embedded material by polymerase chain reaction. J Histochem Cytochem 1991;39:351–4. [DOI] [PubMed] [Google Scholar]
- 6. Inoue T, Nabeshima K, Kataoka H, et al. Feasibility of archival non-buffered formalin-fixed and paraffin-embedded tissues for PCR amplification: an analysis of resected gastric carcinoma. Pathol Int 1996;46:997–1004. [DOI] [PubMed] [Google Scholar]
- 7. Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol 2002;161:1961–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonin S, Petrera F, Niccolini B, et al. PCR analysis in archival postmortem tissues. Mol Pathol 2003;56:184–6. [DOI] [PMC free article] [PubMed] [Google Scholar]