Abstract
Aims: To detect non-viral mRNA in human plasma that has been frozen for three years using a new protocol.
Methods: Plasma from 15 patients with colorectal cancer and 10 normal subjects was separated and frozen with Trizol at −80°C for three years. As a control measure, plasma from 10 of the 15 patients was separated using the same protocol but no Trizol during storage. After three years, all samples were extracted using Trizol and RNeasy before the reverse transcriptase polymerase chain reaction was performed to detect non-viral β catenin mRNA. In addition, extraction of three plasma samples by Trizol or RNeasy independently was carried out for comparison.
Results: β Catenin mRNA was detected in all 15 patient plasma samples and only one of the 10 normal subjects. In contrast, no β catenin mRNA was found in the control and patient samples that were independently extracted by Trizol and RNeasy kit.
Conclusions: This new protocol is a reliable method for extracting non-viral mRNA from the plasma of patients with cancer after longterm storage for three years. Extractions using Trizol and RNeasy kits independently could not isolate mRNA with sufficient quantity and quality for detection.
Keywords: mRNA, plasma, Trizol, β catenin, colorectal cancer
Traditional methods in cancer detection such as colonoscopy and sigmoidoscopy are highly specific and sensitive tests but they are invasive and limited by high costs and demanding expertise.1 Recent reports have indicated that plasma can be an important source of genomic DNA or mRNA in patients with cancer.2,3 Preliminary results on circulating mRNA show that it is even more sensitive than DNA in cancer detection, but is limited by the tedious and inconsistent extraction methods required.2–4 The isolation of non-viral mRNA is most demanding because the quantity is generally much lower than that of viral RNA.5 Viruses actively replicate and enter the bloodstream for dissemination,6 whereas non-viral mRNA, which is subjected to ribonuclease activity in the blood, may be derived from tumour cells as a result of necrosis, apoptosis, or active release.4
“Trizol is a ready to use reagent for the isolation of total RNA, which disrupts cells and dissolves cell components, but at the same time maintains the integrity of RNA”
Preserving the stability of mRNA is a key element to success in clinical research. To investigate whether plasma mRNA is still amplifiable after longterm storage, we devised a new protocol for the manipulation of plasma mRNA that has been frozen for three years, using β catenin mRNA as an example in patients with colorectal cancer.7 This new protocol consists of the use of a combination of Trizol reagent and the RNeasy mini kit. Trizol is a ready to use reagent for the isolation of total RNA from fresh cells or fixed paraffin wax embedded tissues, which disrupts cells and dissolves cell components, but at the same time maintains the integrity of RNA.8 In contrast, the RNeasy kit combines the selective binding properties of a silica gel based membrane with the speed of microspin technology.4 The advantage of this sophisticated technique is that it can isolate all RNA molecules longer than 200 nucleotides, although it has a maximum loading capacity of 100 μg for each mini column.4 In this report, extractions using Trizol reagent and the RNeasy mini kit independently were also evaluated for comparison with the new combined protocol.
METHODS
Our study was approved by the local institutional committee and informed consent was obtained from 15 patients with colorectal cancer and 10 normal subjects in January 1999. For each subject, 6 ml of blood was collected in EDTA coated polypropylene tubes (2 ml each). The tubes were centrifuged at 3838 ×g for eight minutes at 4°C, followed by careful separation of 1 ml plasma from each tube into a ribonuclease free Eppendorf tube with 1 ml Trizol, and stored at −80°C for three years. As a control measure, blood was also collected from 10 of the above patients with colorectal cancer and their plasma was separated and stored at −80°C for three years without Trizol.
After storage for three years, mRNA was extracted using Trizol (Life Technologies, Carlsbad, California, USA), and further purified with the RNeasy kit (Qiagen, Hilden, Germany). In brief, a 2 ml plasma sample with Trizol was mixed with 0.4 ml chloroform before centrifugation. The aqueous phase was collected for mRNA extraction using the RNeasy kit. β Catenin mRNA was detected after treatment with DNase I (Life Technologies) and Omniscript reverse transcriptase (Qiagen), according to the manufacturers’ instructions, and polymerase chain reaction (PCR) assay with a set of intron spanning primers (table 1; primer set 1). To prove the amplifiable nature of those specimens, plasma samples with negative results were subjected to β actin mRNA detection by another set of intron spanning primers (table 1; primer set 2). Aliquots of 6 μl cDNA were used for each PCR reaction, which was carried out using reagents supplied in the GeneAmp DNA amplification kit using AmpliTaq Gold as the polymerase (Perkin-Elmer, Foster City, California, USA). The PCR parameters were as follows: an initial denaturation at 95°C for 10 minutes, followed by 40 cycles of 94°C for 75 seconds, 59°C for 90 seconds, and 72°C for 90 seconds, with a final extension step of 72°C for 10 minutes. PCR products were separated by electrophoresis using a 1.5% agarose gel and visualised by ethidium bromide staining. Duplicate samples were performed for each data point.
Table 1.
Sequences of primers used in our study
| Primer sets | Design | Nucleotide sequence (5′ to 3′) | Product |
| 1 | Intron between exon 3 and 4 of β catenin gene | Sense: TGATTTGATGGAGTTGGACATAntisense: CATTGCATACTGTCCATCAAT | DNA, 450 bpcDNA, 250 bp |
| 2 | Intron between exon 4 and 5 of β actin gene | Sense: AAATCGTGCGTGACATTAAGGAntisense: ATGATGGAGTTGAAGGTAGTT | DNA, 324 bpcDNA, 230 bp |
In addition, patient samples 1 to 3 that had been stored in Trizol were extracted independently using Trizol reagent or the RNeasy mini kit according to the manufacturers’ instructions for comparison. All other steps were the same.
RESULTS
All 15 plasma samples from patients with colorectal cancer showed a 250 bp fragment (fig 1) consistent with β catenin mRNA. To prove that it was derived from mRNA and not DNA, we performed the test on a positive control without DNase I. As expected, it yielded a 250 bp band amplified from mRNA and a 450 bp band amplified from the residual DNA (fig 1). No β catenin mRNA was detected in the negative (plasma from normal subject) and water controls (fig 1).
Figure 1.
Detection of plasma RNA in patients with colorectal cancer: lanes 1–15, β catenin mRNA (250 bp) from 15 patients; lane 16, β catenin mRNA (250 bp) and DNA (450 bp) from a positive control without DNase; lane 17, negative control; lane 18, water control; lane M, molecular markers.
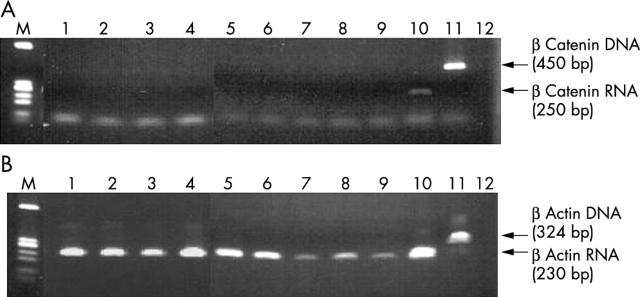
Nine of the 10 plasma samples from normal subjects had no β catenin mRNA (fig 2A). The plasma DNA positive control and the negative control gave the expected results. All samples with negative results were tested for β actin mRNA and all 10 normal subjects showed visible bands of 230 bp (fig 2B). The plasma DNA positive control and the negative control gave the expected results (fig 2B).
Figure 2.
Detection of plasma RNA in normal subjects. (A) Lanes 1–10, β catenin mRNA from 10 normal subjects; lane 11, β catenin DNA from a positive control; lane 12, negative control; lane M, molecular markers. (B) Lanes 1–10, β actin mRNA from 10 normal subjects; lane 11, β actin mRNA from a positive control; lane 12, water control; lane M, molecular markers.
However, β catenin mRNA was not detected in the 10 control colorectal cancer plasma samples that had not had Trizol present during the three year storage period (fig 3A). In addition, the same batch of samples did not contain β actin mRNA (fig 3B).
Figure 3.
Lack of detection of RNA in plasma from patients with colorectal cancer stored for three years without Trizol: (A) lanes 1–10, β catenin mRNA from 10 patients; (B) lanes 1–10, β actin mRNA from 10 patients.
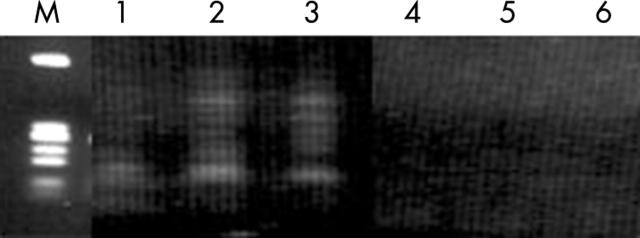
Finally, patient plasma samples 1 to 3 were also extracted independently using either Trizol or the RNeasy kit (fig 4) with the same post-extraction protocols. No β catenin mRNA could be detected by either method.
Figure 4.
Detection of plasma RNA in patients 1–3 with colorectal cancer using both methods independently: lanes 1–3, β catenin mRNA extracted by Trizol; lanes 4–6, β catenin mRNA extracted by RNeasy kit; lane M, molecular markers.
DISCUSSION
The successful demonstration of plasma β catenin mRNA in all patients with colorectal cancer and its absence in normal controls clearly attest to the superiority of our protocol for plasma mRNA extraction after freezing at −80°C for three years. Moreover, our findings also indicate that the addition of Trizol during storage is an important factor in the stabilisation of mRNA over such a long period because both β catenin and β actin mRNA could not be detected using this extraction protocol when Trizol was not present during the three year storage period.
Our protocol, using centrifugation at 3838 ×g for eight minutes, may produce plasma with a rich source of both particle associated and non-particle associated circulating mRNA for detection because cell free plasma requires either a much higher centrifugal force of 99 960 ×g for 120 minutes or a filtration step.9 Therefore, a lower centrifugal force probably produces plasma with more mRNA—Kopreski et al and Silva et al used centrifugal forces as low 830 ×g and 1000 ×g for 10 minutes in their mRNA extraction protocols, respectively.10,11
Take home messages.
We have devised a reliable method using a combination of Trizol and RNeasy kits for extracting non-viral mRNA from the plasma of patients with cancer after longterm storage for three years
Extractions using Trizol and RNeasy kits independently could not isolate mRNA with sufficient quantity and quality for detection
The incidental discovery of β catenin mRNA in the plasma of patients with colorectal cancer but not in most healthy subjects requires further investigation
“Our findings indicate that the addition of Trizol during storage is an important factor in the stabilisation of mRNA over such a long period”
A previous study where mRNA was extracted from 3 ml plasma samples using RNeasy kit and nested PCR had positive rates of only 60% for mammaglobin and 49% for cytokeratin 19.11 Our new protocol increases the percentage of positivity and has the advantage that mRNA can be extracted from only 2 ml of plasma and that it saves time in the first centrifugation step after the addition of one volume of 70% alcohol using the RNeasy kit. In addition, routine rather than nested PCR can be used, which is also an advantage because this last technique is very sensitive but increases the risk of false positivity as a result of contamination. In contrast, we showed that the quality and quantity of extracted mRNA were inadequate for cDNA synthesis when either the Trizol or the RNeasy extraction method was used independently. This might be because although Trizol extracts sufficient mRNA for further processing, the quality is poor, as is the case with many non-specific products. In practice, the Trizol method makes it difficult to obtain a visible and purified mRNA precipitate from plasma for further processing, whereas RNeasy extraction has the disadvantages that the mini spin column has a maximum binding capacity of only 100 μg mRNA and its lytic buffer might form precipitates with plasma that may clot the column, which can greatly reduce the amount of mRNA isolated.
The discovery of β catenin mRNA in the plasma of patients with colorectal cancer but not in most of the healthy subjects is an incidental finding by us and requires further investigation. The only normal subject in whom plasma β catenin mRNA was detected had severe rectal inflammation five years previously and was still suffering from diarrhoea. Selected β catenin cDNA products (seven of 15 colorectal cancer samples) were confirmed by sequencing (data not shown).
In conclusion, this new protocol should be useful because it is the first to isolate high quality non-viral mRNA from patients’ plasma that had been frozen for three years.
Acknowledgments
We thank Dr JKC Chan for valuable discussions. This work was supported by Ho Hung Chiu Medical Foundation Research Grant.
Abbreviations
PCR, polymerase chain reaction
REFERENCES
- 1.Traverso G , Shuber A, Levin B, et al. Detection of APC mutations in fecal DNA from patients with colorectal tumors. N Engl J Med 2002;346:311–20. [DOI] [PubMed] [Google Scholar]
- 2.Lo YM, Johnson PJ. Plasma nucleic acids in the diagnosis and management of malignant disease. Clin Chem 2002;48:1186–93. [PubMed] [Google Scholar]
- 3.Anker P , Stroun Maurice. Progress in the knowledge of circulating nucleic acids: plasma RNA is particle-associated. Can it become a general detection marker for a cancer blood test? Clin Chem 2002;48:1210–11. [PubMed] [Google Scholar]
- 4.Silva JM, Rodriguez R, Garcia JM, et al. Detection of epithelial tumour RNA in the plasma of colon cancer patients is associated with advanced stages and circulating tumour cells. Gut 2002;50:530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stricker RB, Zhu YS, Elswood BF. A simplified method for quantitation of human immunodeficiency virus type 1 (HIV1) RNA in plasma: clinical correlates. Res Virol 1995;146:151–8. [DOI] [PubMed] [Google Scholar]
- 6.Temin HM. The DNA provirus hypothesis: the establishment and implications of RNA-directed DNA synthesis. Science 1976;192:1075. [DOI] [PubMed] [Google Scholar]
- 7.Wong SCC, Lo ESF, Chan AKC, et al. Nuclear β-catenin as a potential prognostic and diagnostic marker in colorectal cancer patients from Hong Kong. Mol Pathol 2003;56:347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shabaana AK, Venkatasubramani R, Narayan NS, et al. Cytokine profiles in paraffin-embedded biopsy samples of lepromatous leprosy patients: semi-quantitative measure of cytokine mRNA using RT-PCR. Int J Lepr Other Mycobact Dis 2001;69:204–14. [PubMed] [Google Scholar]
- 9.Ng EK, Tsui NB, Lam NY, et al. Presence of filterable and nonfilterable mRNA in the plasma of cancer patients and healthy individuals. Clin Chem 2002;48:1212–17. [PubMed] [Google Scholar]
- 10.Kopreski MS, Benko FA, Kwak LW, et al. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res 1999;5:1961–5. [PubMed] [Google Scholar]
- 11.Silva JM, Dominguez G, Silva J, et al. Detection of epithelial messenger RNA in the plasma of breast cancer patients is associated with poor prognosis tumor characteristics. Clin Cancer Res 2001;7:2821–5. [PubMed] [Google Scholar]