Abstract
Background: The amount of argyrophilic nucleolar organiser regions (AgNORs) represents a cell kinetics parameter used in tumour pathology for prognostic purposes. AgNOR expression is directly related to the rate of ribosome biogenesis, which has been recently shown to be controlled also by the tumour suppressor proteins pRb and p53.
Aims: To ascertain the relative prognostic value of AgNORs and of pRb and p53 expression in breast carcinoma.
Methods: This study was carried out on 335 human primary breast carcinomas with a median follow up time of 108 months. AgNORs were measured by morphometric analysis on sections that had been selectively silver stained; expression of p53 and phosphorylated and non-phosphorylated pRb forms was assessed by immunohistochemistry.
Results: Patients were divided into groups with low (249) and high (86) AgNOR values; with normal (267) and mutated p53 (68); and with normal (256) and hyperphosphorylated or deleted pRb (79). Univariate analysis of disease free survival showed that AgNORs and the status of pRb and p53 were significantly related to the patients’ clinical outcome. However, among the four groups characterised by different pRb and p53 status, the AgNOR parameter was not capable of distinguishing subgroups of patients with different clinical outcomes.
Conclusions: These findings indicate that the prognostic value of the AgNOR parameter depends on the status of the tumour suppressor proteins pRb and p53, and it cannot be ascribed to the relation between AgNORs and the cell proliferation rate.
Keywords: argyrophilic ucleolar organiser region breast cancer, p53 pRb prognosis
Nucleolar organiser regions (NORs) are segments of metaphase chromosomes where ribosomal genes are located and which correspond to secondary constrictions.1 During interphase, NORs are located in both the fibrillar centres and the closely associated dense fibrillar component of the nucleolus.2–4 These structures contain all the necessary components for rRNA synthesis and are the sites where the transcription of ribosomal genes occurs.5 A peculiar group of highly argyrophilic acidic proteins is present in the NORs, thus allowing NORs to be clearly and selectively visualised at the light microscopic level by specific silver nitrate staining procedures.6 The silver stained NORs are called AgNORs. AgNORs were introduced into tumour pathology in 1986 by Ploton et al,7 who noted that AgNORs were more abundant in malignant than in benign neoplastic cells. Initially used as a parameter for the diagnosis of malignancy, the AgNOR parameter was found to be more useful for assessing the prognosis of cancer disease.8 More than 300 papers have been published that show a significant correlation between the AgNOR value and patient survival in a large variety of cancers: the greater the AgNOR value, the worse the prognosis.9 The relation between AgNOR values and cancer prognosis was explained on the basis of the biological function of the NORs. In vitro and in vivo studies have shown that the quantity of AgNORs is directly related to the ribosome biogenesis rate, which—in proliferating cells—is tightly related to the length of the cell cycle. The shorter the cell cycle, the greater the synthesis of rRNA for each time unit and, therefore, the quantity of AgNORs present in the nucleolus. Thus, the AgNOR value was thought to be a measure of the rate of cell proliferation.10,11 Because the tumour mass growth rate is one of the most important factors influencing the clinical outcome of cancer, faster growth was suggested to explain the worse prognosis of cancers with high AgNOR values compared with cancers with low AgNOR values.8
“Cancers with changes in pRb and p53 status are generally more aggressive than those with normally functioning pRb and p53 pathways”
There is increasing evidence that the factors controlling cell cycle progression also modulate the rate of ribosome biogenesis. Among these, the most important factors are the tumour suppressor proteins pRb and p53, which are key negative regulators at the G1/S phase restriction point. Both pRb and p53 have been reported to repress the transcription of rRNA genes by affecting the activity of the RNA polymerase I upstream binding factor (UBF).12–16 In most cancers, the mechanisms that control cell cycle progression are perturbed and changes in the pRb and p53 pathways are often responsible for the uncontrolled transit of cancer cells throughout the G1/S phase restriction point. From the clinical point of view, cancers with changes in pRb and p53 status are generally more aggressive than those with normally functioning pRb and p53 pathways.17 pRb deletion or hyperphosphorylation and the mutation of p53 are also associated with enhanced ribosome biogenesis; thus, we wondered whether the worse prognosis of cancers with high AgNOR values resulted either from a high cell proliferation rate or was the consequence of other effects of the altered function of pRb and p53 on cell cycle progression control. Therefore, in our present study we have evaluated the pRb and p53 status and the quantitative distribution of AgNORs in histological sections from 335 human primary breast cancers, to ascertain their relative prognostic value. Visualisation of pRb and p53 was carried out with the use of specific monoclonal antibodies. Quantitative analysis of all parameters was performed by computer assisted image cytometry. Parameters were related to the clinical outcome of the disease as defined by the disease free survival (DFS) interval.
METHODS
Patients
We studied tumour specimens from 335 consecutive cases of primary invasive breast carcinoma collected from 1991 to 1994. Histological diagnosis was performed according to the World Health Organisation criteria. Histological grading was performed in ductal carcinomas according to Elston and Ellis.18 Tumour size was evaluated in freshly obtained tissue, before formalin fixation, and coded according to the UICC pT recommendations. Axillary node status was assessed by pathological staging after axillary node dissection: node metastases were reported as absent (N0) or present (N+). Table 1 reports the histological diagnosis and pT, G, and N status distribution.
Table 1.
Histopathological characteristics of all cases
| Histological characteristic diagnosis | n (%) |
| Histological diagnosis | |
| Ductal carcinoma | 298 (88.9) |
| Lobular carcinoma | 27 (8.1) |
| Pure mucoid carcinoma | 7 (2.1) |
| Sarcomatoid carcinoma | 3 (0.9) |
| Tumour size | |
| T1 | 215 (64.2) |
| T2 | 79 (23.6) |
| T3 | 10 (3) |
| T4 | 31 (9.2) |
| Histological grading | |
| G1 | 25 (8.4) |
| G2 | 214 (71.8) |
| G3 | 59 (19.8) |
| N status | |
| N (0) | 171 (51) |
| N (+) | 164 (49) |
Histological grading was applied to the ductal carcinomas only.
Immunohistochemical assessment
From each case, one block of formalin fixed, paraffin wax embedded tissue was selected, which included a representative tumour area. Serial sections (4 μm thick) were cut, collected on 3-ethoxy-aminoethyl-silane treated slides, and allowed to dry overnight at 37°C.
pRb immunostaining was assessed using two different monoclonal antibodies (MoAbs): the clone G3-245 (BioGenex Laboratories, San Ramon, California, USA), which specifically recognises the phosphorylated pRb form, and the clone 1F8/Rb1 (Neomarkers, Lab Vision, Newmarket, Suffolk, UK), which identifies all forms of pRb. Before immunostaining, sections were microwaved in EDTA buffer solution (pH 8.0) for 10 minutes at 750 W. After cooling down to room temperature, slides were incubated with primary MoAbs overnight at the following dilution: 1/160 for clone G3-245 and 1/30 for clone 1F8/Rb1. The immunostaining reaction was then developed according to the streptavidin–biotin–peroxidase complex method and highlighted using a peroxidase/diaminobenzidine enzymatic reaction.
p53 immunostaining was assessed using an anti-p53 MoAb (clone BP53-12.1; BioGenex Laboratories) which recognises both wild-type and mutated p53. Before immunostaining, sections were microwaved in 10mM citrate buffer solution (pH 6.0) for 17.5 minutes at 750 W. After cooling down to room temperature, slides were incubated with the primary MoAbs overnight at a dilution of 1/1800. The immunostaining reaction was developed and highlighted as described above.
NOR silver staining
AgNOR staining was performed according to the guidelines of the “international committee on AgNOR quantitation”.19 In brief, slides were moved from water to heat resistant plastic Coplin jars, fully immersed in 10mM sodium citrate buffer (pH 6.0), and autoclaved at 120°C for 20 minutes. After cooling down to room temperature in the sodium citrate buffer, slides were stained with silver for 13 minutes at 37°C in the dark by using a solution of one volume of 2% gelatin in 1% aqueous formic acid and two volumes of 50% silver nitrate. Sections were finally dehydrated and mounted in Canada balsam with no counterstaining.
Image cytometry
The quantitative evaluation of all parameters was carried out by image cytometry. p53 and pRb immunostaining was semiquantitatively assessed using the Cytometrica programme (C & V, Bologna, Italy), as detailed previously,20 and expressed as the percentage of labelled nuclear area over the total neoplastic nuclear area in the section (labelling index area; LIa). For each case, at least 2000 cells were evaluated. Morphometric analysis of AgNORs was carried out using the Image-Pro Plus program (Media Cybernetics, Silver Spring, Maryland, USA). The main stages of image processing were as follows: a microscope field was selected by the operator using a ×40 objective lens. The selected image was then captured and stored in the digital memory, and displayed on the colour monitor. Here, the operator interactively defined the grey threshold, which permitted automatic quantification of the black dots corresponding to the silver stained nucleoli. The morphometric analysis was then performed on a cell by cell basis by converging the window over the corresponding nucleus. For each case, the AgNOR area of at least 200 nuclei was measured and the mean (SD) AgNOR area was calculated.
Statistical analysis
Correlations between parameters that are considered to be continuous variables were analysed using the Spearman rank correlation test. Differences between categorical variables were analysed using the ANOVA test. DFS was evaluated by the Kaplan and Meier method, and differences in DFS curves were assessed by the log rank test.21 The best cutoff value for the AgNOR variable was obtained by the receiver operating characteristic curve. Statistical evaluations were performed using the SPSS program package (Statistical Package for Social Science, SPSS Inc, Chicago, Illinois, USA). Values for p less than 0.05 were regarded as significant.
RESULTS
AgNOR quantitative distribution
The silver staining method, originally described by Ploton and colleagues7 and subsequently modified for formalin fixed samples by Öfner et al,22 allows the visualisation of interphase NORs at the light microscopic level as small, darkly stained dots. In our study, the quantitative evaluation of interphase AgNORs was carried out by morphometric analysis, by measuring the area occupied by the silver stained structures. This method is more reliable than directly counting AgNOR numbers because of the difficulty of separating single AgNORs from one another at the light microscopic level.19 In our present morphometric analysis we did not evaluate the spatial distribution of the AgNOR in the nucleolus (that is, the shape of the nucleolus) and its relation to the other biopathological parameters tested. We are aware that, because highly irregularly shaped nucleoli have been reported to represent a cytological parameter of malignancy,23 it might be of interest to carry out a similar investigation. However, such a study would require a very complex and time consuming morphometric analysis,24 which cannot be proposed for routine pathology.
In our series, the mean AgNOR areas ranged from 1.63 to 11.63 μm2, with a mean (SD) value of 4.12 (1.65) μm2 and a median value of 3.71 μm2. In table 2, the relation of AgNOR area to tumour size (pT), histological grade (G), and lymph node status (N) is reported. The AgNOR parameter was found to correlate significantly with both pT and G, whereas no correlation was seen with lymph node status.
Table 2.
Relation between AgNOR area values and pathological parameters
| n | Mean (SD) AgNOR area (μm2) | p Value | |
| Tumour size | |||
| pT1 | 215 | 3.88 (1.49) | 0.001 |
| pT2+pT3+pT4 | 120 | 4.52 (1.82) | |
| Histological grade | |||
| G1 | 25 | 2.95 (0.79) | <0.001 |
| G2 | 214 | 3.94 (1.41) | |
| G3 | 59 | 5.52 (1.92) | |
| N status | |||
| N0 | 171 | 3.94 (1.50) | NS |
| N+ | 164 | 4.29 (1.76) |
AgNOR, argyrophilic nucleolar organiser region; NS, not significant.
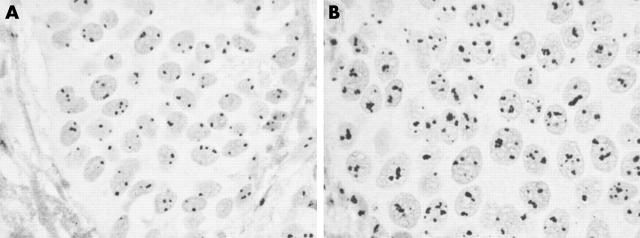
Figure 1 shows two ductal infiltrating carcinomas, which are characterised by different AgNOR quantitative distributions. Very few, solitary, small dots are present in the nuclei of the case shown in fig 1A—each dot corresponds to a small nucleolus. Previous investigations have shown that these nucleoli have a very simple structural organisation, with only one fibrillar centre (ring shaped nucleoli), and low rRNA transcriptional activity.5,6 Figure 1B shows another case in which many silver stained dots are visible within the nuclei. The dots are frequently grouped in large clusters. Each cluster corresponds to a large nucleolus, which contains many fibrillar centres (nucleolonema structured nucleoli) and is very active in ribosome biogenesis.5,6
Figure 1.
Silver stained histological sections of two breast carcinomas. (A) Case with low numbers of argyrophilic nucleolar organiser regions (AgNORs); (B) case with higher numbers of AgNORs.
pRb status
The pRb status was first assessed using an anti-pRb MoAb that specifically recognises the phosphorylated pRb form (clone G3-245). The pRb LIa ranged from 0% to 91.2%, with a mean (SD) value of 14.6% (13.9%) and a median value of 10.3%. Cancers with a pRb LIa > 25% were considered hyperphosphorylated. The 25% cutoff point was chosen because pRb hyperphosphorylation is found mainly in the late G1, S, and G2 phases, whose duration in human cancers is not longer than one quarter of the cell cycle length.25 Therefore, the presence of a pRb LIa > 25 is strongly indicative of an alteration of pRb phosphorylation control.
In 34 cases, a very low positivity for phosphorylated pRb (LIa < 2%) was seen. These cases were thought to include two groups of tumours, namely: (a) tumours in which pRb was present, but phosphorylated only in a few cells; and (b) tumours in which both the pRb forms were absent (probably as a result of pRb deletion). Therefore, these 34 cases were also investigated for the presence of total pRb, using a specific MoAb (clone 1F8/Rb1), which recognises both the phosphorylated and the unphosphorylated/underphosphorylated pRb forms. Ten cases showed positive immunostaining in some cancer cells and the remaining 24 cases showed no immunostaining in the cancer cell population: these last 24 cases were categorised as pRb deleted. Therefore, from the functional point of view, we identified two groups of patients: one group (pRb+), which included both the cases with a presumed deletion of the pRb gene and the cases with high phosphorylated pRb immunostaining (pRb LIa > 25%); the second group (pRb−), which included the cases with low phosphorylated pRb immunostaining (pRb LIa < 25%).
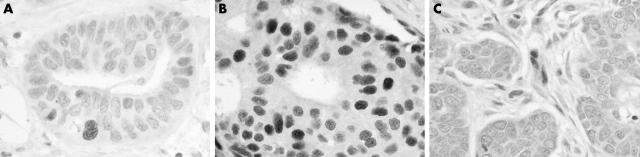
Figure 2A and B shows two infiltrating ductal carcinomas immunostained with the monoclonal antibody specific for the phosphorylated pRb form (clone G3-245): only a few cancer cells are positive in the case shown in fig 2A, whereas a high number of nuclei are positive in the case shown in fig 2B. In both cases, stromal cells are not stained. Figure 2C shows a breast carcinoma with presumed deletion of pRb. When the monoclonal antibody that recognises total pRb (clone 1F8/Rb1) was used the cancer cells did not stain, whereas stromal cells (positive internal controls) were stained.
Figure 2.
pRb immunostaining of three breast carcinomas using two different monoclonal antibodies: (A, B) clone G3-245, which specifically recognises the phosphorylated pRb form, and (C) clone 1F8/Rb1, which reacts with total pRb. (A) Only a few cancer cells are positive in this case but (B) a high number of nuclei are positive in this case. (C) The cancer cells are unstained, indicating a presumable deletion of the pRb gene.
p53 status
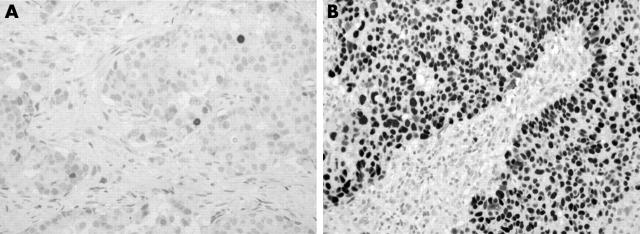
Mutations in the p53 tumour suppressor gene give rise to a protein with a prolonged half life, which can easily be detected by immunohistochemistry because it accumulates in the nuclei. In our series, the percentage of p53 immunostained cells ranged from 0% to 99.7%, with a mean (SD) value of 15.5% (29.1%) and a median value of 2%. For scoring p53 immunoreactivity, we adopted the criteria of Esrig et al,26 who showed that the p53 gene was mutated when 10% or more of the tumour cell nuclei were immunostained. Therefore, we considered samples with at least 10% nuclear reactivity to be p53 mutated (+). Among the 335 cases evaluated, 267 showed a p53 LIa < 10% (p53−) and 68 had a LIa ⩾ 10% (p53+). Figure 3 shows two cancers with a low (fig 3A) and a very high (fig 3B) number of positive cells.
Figure 3.
p53 immunostaining of two breast carcinomas. (A) Only a few cancer cells are positive; (B) p53 accumulation is evident.
AgNOR quantitative distribution in relation to pRb and p53 status
There were significantly more AgNORs in patients with alterations in the tumour suppressor proteins (pRb deleted or hyperphosphorylated, or p53 accumulation) than in those with normal pRb and p53 status (table 3). This was true also when node positive and negative cancers were considered separately (table 4).
Table 3.
Mean AgNOR area according to pRb and p53 status in the whole series of patients
| Groups (n) | Total patients (n = 335) | ||
| Mean (SD) AgNOR area (μm2) | ANOVA analysis (F) | p Value | |
| pRb− (256) | 3.68 (1.35) | 99.88 | 0.000 |
| pRb+ (79) | 5.54 (1.73) | ||
| p53−(267) | 3.79 (1.46) | 59.04 | 0.000 |
| p53+(68) | 5.38 (1.73) | ||
AgNOR, argyrophilic nucleolar organiser region.
Table 4.
Mean AgNOR area according to pRb and p53 status in N0 and N+ subgroups
| Groups (n) | Mean (SD) AgNOR area (μm2) | ANOVA analysis (F) | p Value |
| N0 patients (n = 171) | |||
| pRb− (141) | 3.59 (1.35) | 57.15 | 0.000 |
| pRb+ (30) | 5.58 (1.73) | ||
| p53− (141) | 3.60 (1.22) | 54.35 | 0.000 |
| p53+ (30) | 5.55 (1.69) | 41.27 | |
| N+ patients (n = 164) | |||
| pRb− (115) | 3.77 (1.48) | 0.000 | |
| pRb+ (49) | 5.50 (1.79) | ||
| p53− (126) | 4.00 (1.67) | 15.58 | 0.000 |
| p53+ (38) | 5.24 (1.77) |
AgNOR, argyrophilic nucleolar organiser region.
AgNORs, pRb, and p53 status and cancer prognosis
For statistical analysis, each variable was dichotomised. The AgNOR variable was dichotomised as described earlier. As far as the pRb status is concerned, two groups were considered: one group (pRb+) comprised 24 cases with a presumed deletion of the pRb gene and 55 cases with a phosphorylated pRb LIa > 25% (n = 79; 25.6%), whereas the second group (pRb−) comprised 256 (76.4%) cases with phosphorylated pRb LIa < 25%.
The p53 variable was scored into two groups according to the criteria of Esrig and colleagues23: one group (p53−) comprised 267 (79.7%) tumours with p53 LIa < 10% and a second group (p53+) comprised 68 (20.3%) tumours with p53 LIa ⩾ 10%.
The prognostic significance of the AgNOR, pRb, and p53 variables was tested by univariate DFS analysis, initially on the entire series. All the parameters were found to be significantly correlated with DFS (table 5). Next, the predictive value of the AgNOR parameter in relation to the pRb and p53 status was investigated. For this purpose four groups were defined, characterised by different pRb and p53 status: the first group comprised patients with normal pRb and p53 status (pRb−; p53−); the second group comprised patients with both pRb deletion or hyperexpression and p53 mutation (pRb+; p53+); the third group comprised patients with p53 accumulation and neither pRb deletion nor hyperexpression (p53+; pRb−); whereas the last group comprised patients with either pRb deletion or hyperexpression and no p53 accumulation (pRb+; p53−). The ability of the AgNOR parameter to distinguish two subgroups of patients with different clinical outcome in each of these groups was then evaluated. The AgNOR value could not be used to identify patients with different prognoses in the patient groups defined according the pRb and p53 status (table 6).
Table 5.
DFS analysis of pRb, p53, and AgNOR variables investigated by the log rank test
| Variables | n | % DFS | χ2 | p Value |
| pRb status | ||||
| pRb− | 256 | 74.61 | 21.57 | <0.0001 |
| pRb+ | 79 | 49.37 | ||
| p53 LIa | ||||
| <10% | 267 | 72.66 | 13.30 | 0.0003 |
| ⩾10% | 68 | 52.94 | ||
| AgNOR area | ||||
| <5 μm2 | 249 | 73.49 | 12.64 | 0.0004 |
| >5 μm2 | 86 | 54.65 |
AgNOR, argyrophilic nucleolar organiser region; DFS, disease free survival; LIa, labelling index area.
Table 6.
Predictive impact of the AgNOR variables within subgroups of patients with different pRb/p53 status investigated by the log rank test
| n | % DFS | χ2 | p Value | |
| pRb− p53− | ||||
| AgNOR area <5 μm2 | 203 | 77.83 | 2.39 | 0.1224 |
| AgNOR area >5 μm2 | 26 | 65.38 | ||
| pRb+ p53+ | ||||
| AgNOR area <5 μm2 | 15 | 46.67 | 0 | 0.9735 |
| AgNOR area >5 μm2 | 26 | 50 | ||
| pRb− p53+ | ||||
| AgNOR area <5 μm2 | 13 | 53.85 | 0.32 | 0.5702 |
| AgNOR area >5 μm2 | 14 | 64.29 | ||
| pRb+ p53− | ||||
| AgNOR area <5 μm2 | 18 | 61.11 | 1.52 | 0.2184 |
| AgNOR area >5 μm2 | 20 | 40 |
AgNOR, argyrophilic nucleolar organiser region; DFS, disease free survival.
In a second survival analysis, the groups of patients with node negative and positive cancers were examined separately. Only the pRb status was found to be significantly associated with prognosis in node negative cases, whereas in node positive cases all three parameters were correlated with DFS (table 7). Therefore, the predictive value of the AgNOR parameter in relation to the pRb and p53 status was investigated in node positive cancers only, in the same manner to that used for the entire series. Once again, The AgNOR value could not be used to identify patients with different prognoses in the patient groups defined according the pRb and p53 status (table 8).
Table 7.
DFS analysis of pRb, p53, and AgNOR variables investigated by the log rank test in node negative and node positive tumours
| Variables | Node negative (n = 171) | Node positive (n = 164 ) | ||||||
| n | % DFS | χ2 | p Value | n | % DFS | χ2 | p Value | |
| pRb status | ||||||||
| pRb− | 141 | 84.4 | 6.5 | 0.0108 | 115 | 62.61 | 8.39 | 0.0038 |
| pRb+ | 30 | 66.67 | 49 | 38.78 | ||||
| p53 LIa | ||||||||
| <10% | 141 | 82.98 | 1.71 | 0.1912 | 126 | 61.11 | 10.31 | 0.0013 |
| ⩾10% | 30 | 73.33 | 38 | 36.84 | ||||
| AgNOR area | ||||||||
| <5 μm2 | 135 | 83.7 | 2.53 | 0.1118 | 114 | 61.4 | 6.76 | 0.0093 |
| >5 μm2 | 36 | 72.22 | 50 | 42 | ||||
AgNOR, argyrophilic nucleolar organiser region; DFS, disease free survival; LIa, labelling index area.
Table 8.
Predictive impact of the AgNOR parameter within subgroups of patients with different pRb/p53 status investigated by the log rank test in node positive tumours
| pRb/p53 status | n | % DFS | χ2 | p Value |
| pRb− p53− | ||||
| AgNOR area <5 μm2 | 86 | 68.60 | 1.86 | 0.1726 |
| AgNOR area >5 μm2 | 13 | 53.85 | ||
| pRb+p53+ | ||||
| AgNOR area <5 μm2 | 10 | 40 | 0.43 | 0.5136 |
| AgNOR area >5 μm2 | 12 | 33.33 | ||
| pRb− p53+ | ||||
| AgNOR area <5 μm2 | 7 | 28.57 | 0.55 | 0.4597 |
| AgNOR area >5 μm2 | 9 | 44.44 | ||
| pRb+p53− | ||||
| AgNOR area <5 μm2 | 11 | 45.45 | 0.03 | 0.8558 |
| AgNOR area >5 μm2 | 16 | 37.5 |
AgNOR, argyrophilic nucleolar organiser region; DFS, disease free survival.
DISCUSSION
In our study carried out on 335 cases of human primary breast carcinoma we showed that the quantity of AgNORs and the pRb and the p53 status were prognostic indicators of the clinical outcome of disease. These findings are consistent with those previously reported for AgNORs,9, p53,27,28 and pRb.29–31 In the whole series of breast cancers the three parameters were significantly associated with DFS. However, in node negative cancers, only the pRb variable had significant prognostic value. Our results also showed that, among the four groups characterised by different pRb and p53 status, the AgNOR parameter was not capable of distinguishing subgroups of patients with different clinical outcome, both in the whole series and the node positive group of cancers. Therefore, the prognostic value of the quantitative expression of AgNORs was dependent on the status of the pRb and p53 tumour suppressor proteins.
“The more malignant phenotype of tumours with functional changes in pRb and p53 may be the result of chromosomal instability caused by the loss of restriction point control, rather than an increased cell proliferation rate”
Recent findings on the mechanisms linking the control of cell proliferation to that of cell growth indicate that pRb and p53, in addition to their role in the control of cell cycle progression, also regulate cell growth by modulating the ribosome biogenesis rate. The retinoblastoma protein is a key negative regulator at the G1 phase restriction point by binding the E2F family of transcription factors, thus preventing their activity. The E2F factors stimulate both the expression of a series of genes involved in overriding the restriction point and progression throughout the S phase.32 Phosphorylation by cyclin dependent kinase complexes inactivates the suppressive role of pRb,33 by disrupting its association with the E2F factors. There is evidence that pRb also modulates the synthesis of rRNA.12 It was shown that pRb acts as a transcriptional repressor by interfering with the assembly of transcription initiation complexes. The inhibitory effect on RNA polymerase I transcription was the consequence of the binding of hypophosphorylated pRb to UBF.13,14,34 During the cell cycle the sequential activation of the cyclin dependent kinases progressively increases the degree of pRb phosphorylation from early G1 phase, during which it is hypophosphorylated, to G2 phase, when pRb is hyperphosphorylated.35 The synthesis of rRNA progressively increases from the G1 to G2 phase accordingly.12–14 The other major tumour suppressor, p53, is a transcription factor induced by either DNA damage or inappropriate mitogenic signalling, which induces cell cycle arrest at the G1 phase or apoptosis.36 Wild-type, but not mutant, p53 hinders RNA polymerase I transcription by interacting with SL1, thus impairing the formation of the initiation complex. Therefore, p53 deficient cells display increased rRNA synthesis.15,16 Consistent with these observations, we found that tumours characterised by hyperphosphorylated or deleted pRb and/or p53 mutation have a greater AgNOR value than those with a normal pRb and p53 status. In fact, by modulating the transcriptional activity of RNA polymerase I, pRb and p53 are responsible for the different levels of expression of AgNORs, which reflect the rate of ribosome biogenesis. In previous studies we have shown that the quantity of AgNORs and the expression of the major nucleolar proteins involved in the control of rRNA synthesis and processing—such as UBF, topoisomerase I, nucleolin, fibrillarin, and protein B23—are tightly and directly related to RNA polymerase I transcriptional activity.10 We have also shown that both the quantity of AgNORs and the ribosome biogenesis rate are related to the cell doubling time and to the tumour growth rate: the greater the quantity of AgNORs, the faster the cell proliferation and the neoplastic mass expansion.10,11 Thus, we suggested that the association of the quantity of AgNORs with cancer prognosis was a consequence of the fact that the AgNOR value is a measure of the rate of cell proliferation, which, together with the number of proliferating cells, determines the tumour mass growth rate. In other words, tumours with high AgNOR values were characterised by a worse prognosis than those with low AgNOR values because they were growing more rapidly.8 Our present findings, which show a tight association between alterations in pRb and p53 status and high AgNOR values, suggest that the worse prognosis seen in patients with changes in the tumour suppressor proteins might be related to their effect on ribosome biogenesis and, consequently, on the rate of cell proliferation. However, in cancers with normal pRb and p53 status, the prognosis of patients with high AgNOR values was not significantly different from that of patients with low AgNOR values, indicating that the predictive significance of AgNORs cannot be ascribed to their effects on ribosome biogenesis and the cell proliferation rate. Probably, the more malignant phenotype of tumours with functional changes in pRb and p53 may be the result of chromosomal instability caused by the loss of restriction point control, rather than an increased cell proliferation rate.
Take home messages.
In a series of breast carcinomas, univariate analysis of disease free survival showed that argyrophilic nucleolar organiser regions (AgNORs) and the status of pRb and p53 were significantly related to the patients’ clinical outcome
However, among the four groups characterised by different pRb and p53 status, the AgNOR parameter could not distinguish the subgroups of patients with different clinical outcome
Thus, the prognostic value of the AgNOR parameter depends on the status of the tumour suppressor proteins pRb and p53, and cannot be ascribed to the relation between AgNORs and the cell proliferation rate
Nonetheless, the AgNOR value is still a useful prognostic indicator in a variety of human cancers and the measurement of AgNORs should still be recommended in routine pathology for prognostic purposes given its rapid execution, simplicity, and low cost
These findings do not reduce the importance of the AgNOR parameter in tumour pathology. Even if the prognostic value of AgNORs is not a result of them being indicators of the cell proliferation rate, there is no doubt that the AgNOR value remains a strong prognostic indicator in a variety of human cancers.9 For this reason, we feel that the measurement of AgNORs should still be recommended in routine pathology for prognostic purposes given its rapid execution, simplicity, and low cost.
Acknowledgments
This work was supported by Grants from University of Bologna and Pallotti’s Legacy for Cancer Research.
Abbreviations
AgNOR, argyrophilic nucleolar organiser region
DFS, disease free survival
LIa, labelling index area
MoAb, monoclonal antibody
NOR, nucleolar organiser region
UBF, upstream binding factor
REFERENCES
- 1.Howell W M. Selective staining of nucleolus organiser regions (NORs). In: Busch H, Rothblum L, eds. The cell nucleus. New York: Academic Press, 1982:89–143.
- 2.Bourgeois C A, Hernandez-Verdun D, Hubert J, et al. Silver staining of NORs in electron microscopy. Exp Cell Res 1979;123:449–52. [DOI] [PubMed] [Google Scholar]
- 3.Hernandez-Verdun D , Hubert J, Bourgeois C A, et al. Ultrastructural localization of Ag-NOR stained proteins in the nucleolus during the cell cycle and in other nucleolar structures. Chromosoma 1980;79:349–62. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez-Verdun D . Structural organization of the nucleolus in mammalian cells. Methods Achiev Exp Pathol 1986;12:26–62. [PubMed] [Google Scholar]
- 5.Derenzini M , Thiry M, Goessens G. Ultrastructural cytochemistry of the mammalian cell nucleolus. J Histochem Cytochem 1990;38:1237–56. [DOI] [PubMed] [Google Scholar]
- 6.Derenzini M , Ploton D. Interphase nucleolar organiser regions in cancer cells. Int Rev Exp Pathol 1991;32:149–92. [DOI] [PubMed] [Google Scholar]
- 7.Ploton D , Menager M, Jeannesson P, et al. Improvement in the staining and in the visualization of the argyrophilic proteins of the nucleolar organiser regions at the optical level. Histochem J 1986;8:5–14. [DOI] [PubMed] [Google Scholar]
- 8.Derenzini M . The AgNORs. Micron 2000;31:117–20. [DOI] [PubMed] [Google Scholar]
- 9.Pich A , Chiusa L, Margaria E. Prognostic relevance of AgNORs in tumour pathology. Micron 2000;31:133–41. [DOI] [PubMed] [Google Scholar]
- 10.Derenzini M , Treré D, Pession A, et al. Nucleolar function and size in cancer cells. Am J Pathol 1998;152:1291–7. [PMC free article] [PubMed] [Google Scholar]
- 11.Derenzini M , Trere D, Pession A, et al. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J Pathol 2000;191:181–6. [DOI] [PubMed] [Google Scholar]
- 12.Cavanaugh A H, Hempel W M, Taylor L J, et al. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 1995;374:177–80. [DOI] [PubMed] [Google Scholar]
- 13.Voit R , Schäfer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol Cell Biol 1997;17:4230–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannan K M, Kennedy B K, Cavanaugh A H, et al. RNA polymerase I transcription in confluent cells: Rb downregulates rDNA transcription during confluence-induced cell cycle arrest. Oncogene 2000;19:3487–97. [DOI] [PubMed] [Google Scholar]
- 15.Budde A , Grummt I. p53 represses ribosomal gene transcription. Oncogene 1999;18:1119–24. [DOI] [PubMed] [Google Scholar]
- 16.Zhai W , Comai L. Repression of RNA polymerase I transcription by the tumour suppressor p53. Mol Cell Biol 2000;20:5930–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cordon-Cardo C . Mutations of cell cycle regulators. Biological and clinical implications for human neoplasia. Am J Pathol 1995;147:545–60. [PMC free article] [PubMed] [Google Scholar]
- 18.Elston C W, Ellis I O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 1991;19:403–10. [DOI] [PubMed] [Google Scholar]
- 19.Treré D . AgNOR staining and quantification. Micron 2000;31:127–31. [DOI] [PubMed] [Google Scholar]
- 20.Faccioli S , Chieco P, Gramantieri L, et al. Cytometric measurement of cell proliferation in echo-guided biopsies from focal lesions of the liver. Mod Pathol 1996;9:120–5. [PubMed] [Google Scholar]
- 21.Altman D G, De Stavola B L, Love S B, et al. Review of survival analyses published in cancer journals. Br J Cancer 1995;72:511–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Öfner D , Bankfalvi A, Rieheman K, et al. Wet autoclave pretreatment improves the visualization of silver-stained nucleolar organiser region-associated proteins in routinely formalin-fixed and paraffin-embedded tissues. Mod Pathol 1994;7:946–50. [PubMed] [Google Scholar]
- 23.Busch H , Smetana K. Nucleoli of tumor cells. In: Busch H, Smetana K, eds. The nucleolus. New York: Academic Press, 1970:448–71.
- 24.Hufnagl P , Guski H, Schulz H J. Measuring of AgNORs using image analysis. Zentralbl Pathol 1994;14:31–5. [PubMed] [Google Scholar]
- 25.Rew D A, Wilson G D. Cell production rates in human tissues and tumours and their significance. Part II: clinical data. Eur J Surg Oncol 2000;26:405–17. [DOI] [PubMed] [Google Scholar]
- 26.Esrig D , Spruck C H3rd, Nichols P W, et al. 53 nuclear protein accumulation correlates with mutations in the p53 gene, tumour grade, and stage in bladder cancer. Am J Pathol 1993;143:1389–97. [PMC free article] [PubMed] [Google Scholar]
- 27.Bergh J . Clinical studies of p53 in treatment and benefit of breast cancer patients. Endocr Relat Cancer 1999;6:51–9. [DOI] [PubMed] [Google Scholar]
- 28.Borresen-Dale A L. TP53 and breast cancer. Hum Mutat 2003;21:292–300. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen N H, Loden M, Cajander J, et al. G1-S transition defects occur in most breast cancers and predict outcome. Breast Cancer Res Treat 1999;56:105–12. [DOI] [PubMed] [Google Scholar]
- 30.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell 1995;81:323–30. [DOI] [PubMed] [Google Scholar]
- 31.Wakasugi E , Kobayashi T, Tamaki Y, et al. Analysis of phosphorylation of pRB and its regulatory proteins in breast cancer. J Clin Pathol 1997;50:407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harbour J , Dean D. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev 2000;14:2393–409. [DOI] [PubMed] [Google Scholar]
- 33.Ezhevsky S , Ho A, Becker-Hapak M, et al. Differential regulation of retinoblastoma tumour suppressor protein by G1 cyclin-dependent kinase complexes in vivo. Mol Cell Biol 2001;21:4773–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannan K M, Hannan R D, Smith S D, et al. Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene 2000;19:4988–99. [DOI] [PubMed] [Google Scholar]
- 35.Donjerkovic D , Scott D W. Regulation of the G1 phase of the mammalian cell cycle. Cell Res 2000;10:1–16. [DOI] [PubMed] [Google Scholar]
- 36.Levine A J. p53, the cellular gatekeeper for growth and division. Cell 1997;88:323–31. [DOI] [PubMed] [Google Scholar]