Abstract
The mammalian adenosine deaminases acting on RNA (ADARs) constitute a family of sequence-related proteins involved in pre-mRNA editing of nuclear transcripts through site-specific adenosine modification. We report here the identification and characterization of a human ADAR protein, hADAT1, that specifically deaminates adenosine 37 to inosine in eukaryotic tRNAAla. It represents the functional homologue of the recently identified yeast protein Tad1p [Gerber, A., Grosjean, H., Melcher, T. & Keller, W. (1998) EMBO J. 17, 4780–4789]. The hADAT1 cDNA predicts a protein of 502 aa whose sequence displays strongest overall homology to a Drosophila melanogaster ORF (50% similarity, 32% identity), and the catalytic domain is closely related to the other ADAR proteins. In vitro, the recombinantly expressed and purified hADAT1 protein efficiently and specifically deaminates A37 in the anticodon loop of tRNAAla from higher eukaryotes and with lower efficiency from lower eukaryotes. It does not modify adenosines residing in double-stranded RNA or in pre-mRNAs that serve as substrates for ADAR1 or ADAR2. The anticodon stem–loop of tRNAAla alone is not a functional substrate for hADAT1. The enzyme is expressed ubiquitously in human tissues and is represented by a single gene. The identification and cloning of hADAT1 should help to elucidate the physiological significance of this unique modification in tRNAAla, which is conserved from yeast to man.
The family of mammalian adenosine deaminases acting on RNA (ADARs) deaminates adenosine (A) to produce inosine (I) at specific sites (reviewed in refs. 1–3). A growing number of nuclear pre-mRNAs have been found to be substrates for modification by ADARs, leading to transcripts with single or multiple codon changes in the respective mRNA sequence (1, 3, 4). Because inosine prefers to base pair with cytidine, it is read as guanosine by the translational machinery (5). The corresponding proteins have amino acid substitutions that often lead to significantly altered biological function (6, 7). Even though only a small number of RNA-editing substrates have been identified, the ubiquitous expression pattern of the candidate editases (8, 9) as well as the substantial amount of inosine found in mRNA from mammalian tissue isolates (10) indicate that adenosine deamination, together with other types of RNA editing, represents an important mechanism for increasing functional diversity in eukaryotes. ADARs are not active on adenosine-mononucleotides and are only distantly related by sequence to adenosine and AMP deaminases involved in purine metabolism; however, they use the same reaction mechanism of hydrolytic deamination for the conversion of A to I (11–13). The family of mammalian ADAR proteins currently comprise the three cloned enzymes ADAR1 (14–16), ADAR2 (9, 17, 18), and RED2 (19). ADAR1 and ADAR2 have been shown to recognize tertiary-structure elements in their partially double-stranded (ds) RNA substrates formed through base pairing between exonic and intronic sequences (20). In these computer-predicted secondary RNA structures, the targeted adenosine is either base paired, mismatched, or contained in a loop (4, 20–22). In vitro, ADAR1 and ADAR2 display different but overlapping activities on characterized editing substrates (4, 9). The conserved catalytic deaminase domain and two (ADAR2, RED2) or three (ADAR1) dsRNA-binding motifs are the common structural features of all mammalian ADAR proteins.
Although the existence of inosine in nuclear mRNA was only established recently (10) and predicted from the evidence of pre-mRNA editing substrates targeted by adenosine deaminases, it has long been known to occur in tRNAs (23). In higher eukaryotes, eight tRNAs (seven in yeast) contain inosine in the first anticodon position (I34), and N1-methylinosine is found exclusively in tRNAAla at position 37, 3′ adjacent to the anticodon (reviewed in ref. 24; see also ref. 25). In studies with partially purified deaminase activities, it has been shown that the reaction mechanism for the formation of inosine in tRNAs also proceeds via hydrolytic deamination of adenosine and that I34 and I37 are generated by different enzymatic machinery (24, 26). In a second step, I37 is further modified to N1-methylinosine by a separate enzymatic activity (24). Recently, the yeast protein Tad1p/scADAT1 was identified by sequence homology to the catalytic deaminase domain of ADAR proteins (27). Tad1p specifically deaminates A37 in the anticodon loop of eukaryotic tRNAAla, suggesting an evolutionary link between tRNA modification and pre-mRNA editing. Modification of A37 by Tad1p, which lacks any known RNA-binding motif, was strictly dependent on the correct folding of the tRNAAla substrate as well as the local conformation of the anticodon loop (27).
We have now isolated a human cDNA that has a sequence related to the family of ADAR enzymes. The recombinant protein specifically modifies A37 in tRNAAla from higher eukaryotes (human and Bombyx mori) with high efficiency in vitro. With lower efficiency, it complements for Tad1p activity on yeast tRNAAla in vitro and does not crossreact with substrates of other ADAR family members. Thus, hADAT1 (human adenosine deaminase acting on tRNA) probably represents the human counterpart of the yeast protein Tad1p.
MATERIALS AND METHODS
Databases and Programs.
The GenBank and the database of expressed sequence tags (dbest) were searched by using the Gapped Basic Local Alignment Tool (BLAST version 2.0; ref. 28). Sequence data for Trypanosoma brucei was obtained from the Sanger Centre website at http://www.sanger.ac.uk/Projects/T_brucei, and sequence data for Candida albicans was obtained from the Stanford DNA Sequencing and Technology website at http://www-sequence.stanford.edu/group/candida. Multiple alignments of amino acid sequences were performed with clustalw (version 1.73) by using a blosum matrix.
Isolation and Analysis of ADAT1 Phage Clones.
A human brain cDNA library in bacteriophage λgt10 (CLONTECH) was screened initially by hybridization with a 150-bp 32P-labeled DNA probe obtained by PCR using specific primers AA16U and AA16D deduced from a human expressed sequence tag (GenBank accession no. AA161179) with a human lung cDNA library (CLONTECH) as a template. After an additional round of hybridization and screening with a 400-bp cDNA probe derived from the 5′-end of clone λ18, four independent and overlapping λ phage clones were isolated. They were analyzed by restriction mapping using standard protocols (29), and two of them were sequenced completely. DNA sequencing was done either by the dideoxy method (Sequenase 2.0, United States Biochemicals), or by the Massachusetts Institute of Technology Cancer Center Sequencing Facility. For confirmation of the assembled ADAT cDNA sequence in the 5′-end, a rapid amplification of cDNA ends experiment was performed by using total RNA from human embryonic kidney cells (HEK293; ATCC CRL 1573). Total RNA was prepared by using TRIzol-reagent (GIBCO/BRL) according to the manufacturer’s protocol. Independent reverse transcription reactions with Superscript reverse transcriptase (GIBCO/BRL) were performed with antisense primers A5U and A6U at 45°C for 1 h. The resulting cDNA products were 3′ tailed with oligo(dA) by using Terminal Deoxyribonucleotidyltransferase (Boehringer Mannheim). Primers for two successive rounds of PCR were PCR4U and A6U (first PCR) or A7U (second PCR), respectively, and 5′-specific PCR amplicons were analyzed for their sequence.
Northern Blot.
A human multiple-tissue Northern blot (CLONTECH) was hybridized with a 1.5-kilobase (kb) 32P-labeled cDNA probe covering the complete hADAT1 ORF. After hybridization in ExpressHyb hybridization solution (CLONTECH) for 1 hr at 68°C, the blot was washed for 30 min at room temperature in solutions containing 0.1% SDS and 2×, 1×, 0.5×, and 0.2× SSC, successively and then in 0.1× SSC/0.1% SDS at 55°C for 45 min. The filter was exposed to Kodak X-Omat film for 14 days at −70°C. Quantitative signal analysis was done by PhosphorImaging (Molecular Dynamics). As a control, the expression levels of human β-actin were determined by hybridization of the same filter with a specific 1.8-kb β-actin cDNA probe under identical conditions for hybridization and washing as described above.
DNA Constructs and Oligonucleotide Primers.
The mammalian expression vector pEGFP (CLONTECH) with a deleted enhanced green fluorescent protein (EGFP) coding region, was modified to include a Flag epitope coding sequence upstream of the multiple cloning site as well as six His codons downstream and is referred to as pExFH. Cloned full-length cDNA for human ADAR1 (14) and rat ADAR2 (9) were inserted in-frame, yielding an N-terminal Flag epitope for the expressed proteins. Human ADAR1 cDNA was transferred in multiple cloning steps including cDNA fragments as well as PCR-amplified N- and C-terminal regions. The rat ADAR2 coding sequence was PCR-amplified from its source vector and cloned into the BglII and EcoRI sites of the pExFH vector. The human ADAT1 ORF was inserted into the BglII and EcoRI sites of pExFH after PCR amplification with primers A10D and A9U. The correct sequence of the ORF was confirmed for all expression constructs. Single point mutations in hADAT1 cDNA were introduced with the QuikChange site-directed mutagenesis kit (Stratagene).
The chimeras ADAR2/ADAT1 (rADAR2 amino acid residues 1–306 fused to hADAT1 residues 1–502), ADAR2/ΔADAT1 (rADAR2 amino acid residues 1–400 fused to hADAT1 residues 50–502), and ADAR2/ADAR1 (rADAR2 residues 1–306 fused to hADAR1 residues 808-1226) were constructed by PCR-mediated mutagenesis.
For baculovirus-mediated expression of the various native proteins and chimeras in insect cells, the respective NheI–SphI fragment encompassing the ORF and 1 kb of downstream vector sequence of each pExFH construct was ligated to a XbaI–SphI-cut pFastBac1 donor plasmid vector (GIBCO/BRL).
hADAT1 specific oligonucleotide primers were AA16D, 5′-CGCCGGTACCGTGGA-GACAGAACACGCTCC-3′; AA16U, 5′-CGCGAATTCCCAATGACCACAGCTG-ACAG-3′; A5U, 5′-AGTGTTCATAGCATAGCTGAG-3′; A6U, 5′-CGGTCCACAT-GGTCTGAGCTG-3′; A7U, 5′-CGTCTAGATCTAGGGAGGCATCACAAGGC-3′; PCR4U, 5′-GACACGGTACCACACAACGG-3′; A10D, 5′-GGAGAAGATCTT-GGACCGCGGATGAGATTGC-3′; and A9U, 5′-CGCGAATTCCTTGAACTGG-TGATAATCCGG-3′.
Cell Culture and Transfections.
For production of recombinant proteins in insect cells the expression constructs were transfected into DH10BAC cells (GIBCO/BRL) and the recombinant bacmid DNAs were isolated. Sf9 insect cells (ATCC CRL 1711) were seeded onto six-well tissue culture plates for small scale or transferred to 250-ml spinner culture flasks for large-scale protein expression. The CaCl2-mediated transfection of bacmid DNA was performed with PharMingen transfection buffers A and B according to the manufacture’s protocol. Preparation of cellular extracts and Western blots were performed as described (4).
Purification of Recombinant Protein.
Recombinant wild-type and mutant hADAT1 was purified from cellular extracts of baculovirus-infected Sf9 cells through affinity chromatography with an Anti-Flag M2 affinity gel matrix (Sigma) at 4°C. The cellular extract from ≈5 × 107 cells was diluted 1:10 in TBS buffer (50 mM Tris/HCl, pH 7.5/150 mM NaCl/1 mM DTT) and applied to 0.3 ml of affinity resin preequilibrated with TBS. After washing with 15 vol of TBS, the bound protein was eluted with 200 μg/ml Flag-peptide (Sigma) in TBS. Aliquots from all purification steps were separated on SDS/PAGE gels, and proteins were visualized by Coomassie staining, or the recombinant protein was detected by immunoblot with a mouse anti-Flag M2 monoclonal antibody (Eastman Kodak) after transfer of the proteins to nitrocellulose as described (4).
tRNA Substrates and Enzyme Assays.
The human tRNAAla gene was deduced from the tRNAAla transcript sequence in the database (no. RA9990) (25), harboring I34 and m1I37, because the corresponding human genomic sequence is not available. The gene was prepared in two consecutive steps by hybridization and ligation of complementary and partially overlapping oligonucleotides into the EcoRI and HindIII sites of pGEM (Promega) plasmid vector carrying an upstream T7 RNA polymerase promoter. All other tRNA genes were constructed as described (27). Transcription of tRNA genes and in vitro tRNA-specific deaminase assays were performed as described (27) by using ≈20 ng of recombinant, purified hADAT1 and 200 fmol of 33P-labeled substrate. Yeast strains and methods were as described (27).
RESULTS
Isolation and Characterization of Human ADAT1 cDNA Clones.
A hallmark feature of the ADAR family of RNA editing enzymes is a number of highly conserved sequence motifs within their catalytic domain including the residues involved in putative Zn2+ coordination and proton shuttling. In a database search with sections of the ADAR1, ADAR2, and RED2 amino acid sequences from the deaminase region, we noticed a human expressed sequence tag (accession no. AA161179) with high sequence identity to a conserved stretch of amino acids in the 3′ end of the deaminase domain (including the motifs MSCSDK… QGALL… PIY). Screening of a human brain cDNA library yielded four independent and partially overlapping cDNA clones. Sequencing revealed an ORF of 1,509 bp predicting a polypeptide of 502 aa with a calculated molecular mass of 55.3 kDa. The cDNA sequence harbors an in-frame stop codon preceding the predicted ATG translational start codon, which resides in a consensus Kozak sequence motif (30). An additional 159 nt of 5′ untranslated sequence with in-frame stop codons in all reading frames is present on the longest cDNA clone. A 5′-rapid amplification of cDNA ends experiment was performed on total RNA from human embryonic kidney cells (HEK293), confirming these findings.
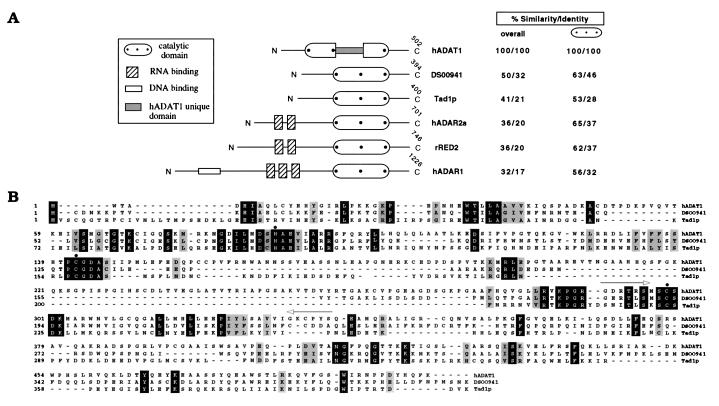
The deduced amino acid sequence of hADAT1 is 32% identical (50% similar) to a region of a Drosophila melanogaster genomic sequence in the database (GenBank accession no. AC001659; Fig. 1). Within the deaminase region, hADAT1 displays high sequence similarity (>50%) with the mammalian pre-mRNA-editing enzymes (ADAR1, ADAR2, RED2) and the yeast tRNA deaminase Tad1p (Fig. 1). No dsRNA-binding motif or any other known RNA interaction domain is present, whereas a unique stretch of 90 aa resides within the deaminase catalytic domain that does not resemble any sequence in the databases. A Southern blot analysis of human genomic DNA revealed that hADAT1 is represented by a single gene (data not shown).
Figure 1.
Domain architecture and amino acid sequence of hADAT1 compared with other ADAR proteins. (A) Schematic representation of functional domains. The percent values for sequence similarity and identity throughout the complete ORF (overall) or within the deaminase domain between hADAT1 and individual deaminases are indicated. DS00941, Drosophila ORF deduced from genomic sequence of chromosome 2 (GenBank accession no. AC001659), nucleotides 71,207–72,078 and 72,136–72,448, Tad1p (AJ007297), hADAR2a (U82120), rRED2 (U74586), and hADAR1 (U10439). The positions of hADAT1-specific PCR primers AA16D and AA16U are depicted by arrows. (B) The deduced amino acid sequence of hADAT1 aligned with the two ADARs displaying the highest overall similarity. Identical residues are highlighted in black, positions with conserved similar amino acids in gray. Three putative Zn2+-chelating residues are marked by a ⋅.
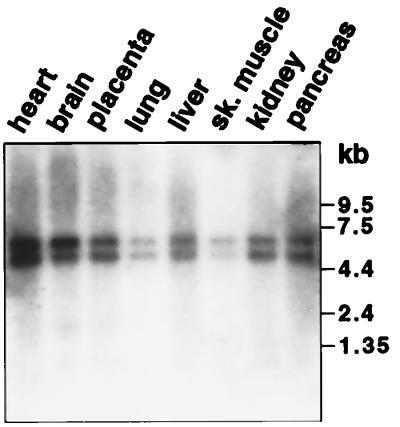
We analyzed the mRNA expression of ADAT1 in human tissues by Northern hybridization with a 1.5-kb cDNA probe encompassing its complete ORF (Fig. 2). Two transcripts, ≈5 kb and ≈6.5 kb in length, respectively, were detected in all tissues, with highest expression levels in heart, brain, and pancreas and lowest levels in lung and skeletal muscle. The relative signal intensities of the two transcripts were approximately equal in all tissues.
Figure 2.
Expression of ADAT1 mRNA in human tissues. A northern blot containing 2 μg of human poly(A)+ RNAs was hybridized with a 1.5-kb hADAT cDNA probe. Size markers in kb are shown on the right. Reprobing of the blot with a β-actin cDNA probe (data not shown) indicated that relative RNA amounts in lanes differed by no more than three-fold.
Expression of Recombinant hADAT1 in Insect Cells.
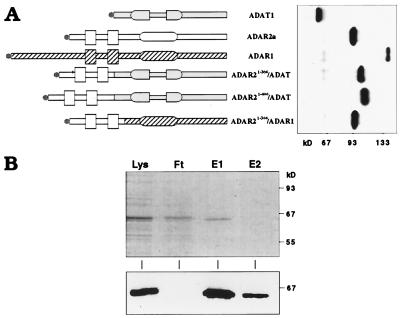
The 1.5-kb ORF of hADAT1 engineered to carry an N-terminal Flag epitope was overexpressed in Sf9 insect cells through infection with recombinant baculovirus. As judged by immunoblot analysis, the expressed protein was of the expected size. However, reverse transcription–PCR and sequencing of hADAT1-specific mRNA from infected insect cells revealed the occurrence of an internal in-frame splicing event that quantitatively removes 40 aa (residues 99–138) of the catalytic region (data not shown). Reverse transcription–PCR analysis on cDNA from human brain and lung (data not shown) demonstrated that this does not represent a natural case of alternative splicing but is likely an aberrant event related to the use of intronless cDNA (31). By using site-directed mutagenesis, three silent mutations (CAG → GTC) were introduced that eliminated internal splicing of the cDNA resulting in full-length expression of hADAT1 (Fig. 3). This construct was used in all subsequent experiments. Wild-type hADAT1, as well as a point mutant changing the conserved sequence motif HAE to QAD in the deaminase domain [known to abolish catalytic function in ADAR1 (4)], was overexpressed and purified by affinity chromatography (>95% pure as judged by Coomassie stain; Fig. 3) for use in functional assays.
Figure 3.
Expression and purification of recombinant hADAT1. (A) Domain structures (symbols as in Fig. 1A) and Western blot analysis of native and chimeric proteins expressed in Sf9 insect cells. Gray, open or hatched areas, respectively, denote ADAT1, ADAR2a, and ADAR1 sequences. The N-terminal Flag epitope is indicated by ●, and size markers are shown on the bottom. (B) Overexpressed hADAT1 was purified by using an anti-Flag-affinity gel matrix, and aliquots from purification steps were separated on SDS/PAGE and analyzed by Coomassie staining (Upper) and Western blotting with M2 anti-Flag mAb (Lower). Lys, lysate; Ft, flowthrough; E1/E2, elution fractions 1 and 2. Size markers are shown on the right.
HADAT1 Specifically Deaminates A37 to I37 in tRNA Ala.
The high sequence homology between the catalytic deaminase domains of hADAT1 and the other ADAR proteins strongly suggested adenosine deamination as the enzymatic function of hADAT1. Because of the lack of any known substrate-interaction domains, we first tested the ability of hADAT1 to target adenosines in characterized substrates of other ADAR proteins, such as extended dsRNA molecules and minigene transcripts of glutamate receptor pre-mRNAs. No inosine could be detected after incubation of purified hADAT1 protein with any of these [32P]adenosine-labeled substrates in vitro, nor did the addition of purified, recombinant hADAT1 to cellular extracts from HEK293 cells increase the level of inosine formation in these substrates above endogenous levels (data not shown). Chimeric proteins fusing the dsRNA binding domains of ADAR2 to either full-length hADAT1 or an N-terminal deletion mutant of hADAT1 (Fig. 3) were constructed and tested the same way. Whereas the analog fusion-construct of the ADAR1 catalytic domain and the ADAR2 dsRNA-binding motifs (Fig. 3) resulted in a protein with substantial adenosine deaminase activity on extended dsRNA, the ADAR2/ADAT1 chimeras were inactive (data not shown; see also ref. 19).
Because inosine has been detected in eukaryotic tRNAs and (given their L-shaped architecture) they represent a structurally distinct class of RNA molecules, we tested hADAT1 for adenosine deaminase activity on in vitro-transcribed tRNAAla from Saccharomyces cereviseae (lower eukaryote) and B. mori (higher eukaryote). The B. mori tRNAAla is 90% identical in nucleotide sequence and 100% identical in the anticodon stem–loop with human tRNAAla, and it was found to be a functional substrate for the A34- and A37-specific deaminase activities from rat and rabbit liver extracts (24).
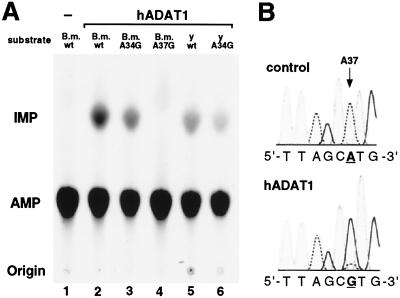
Our results from incubations of recombinant, affinity-purified hADAT1 with in vitro-transcribed and [33P]adenosine-labeled wild-type as well as mutant tRNA substrates (Fig. 4A) unambiguously show that hADAT1 catalyzes the conversion of A37 to I37 specifically and with high efficiency. No inosine is generated in a mutant B. mori tRNAAla substrate with A37 substituted by G, whereas an A34 → G34 mutation did not alter the amount of inosine produced (Fig. 4A).
Figure 4.
(A) tRNA-specific adenosine deaminase assay. 33P-labeled alanine tRNAs (200 fmol) from B. mori (B.m.) and yeast (y) were incubated for 1 h with purified, recombinant hADAT1 and analyzed by one-dimensional TLC after P1 nuclease digestion of the processed RNA. Lane 1, no protein; lanes 2–6, 1.5 μl of hADAT1. A34G, mutant tRNA containing G34 instead of A34; A37G, tRNA with A37 to G37 mutation. The spots at the bottom are the origin; the spots in the middle correspond to AMP; the spots at the top correspond to IMP. (B) Sequence analysis of in vitro-assayed human tRNAAla. In vitro-transcribed human tRNAAla was incubated with 2 μl of recombinant, purified human ADAT1 (hADAT1) or with buffer (control). Human tRNAAla-specific reverse transcription–PCR products were sequenced. Only the anticodon loop-region is shown, and nucleotide position 37 is underlined. Adenosine peaks are indicated by a dashed line, guanosine peaks are bold. Because I prefers to base pair with C, I is represented as G.
Quantification by PhosphorImaging demonstrated that the efficiency of adenosine conversion by recombinant hADAT1 is higher for tRNAAla from B. mori (0.83 mol I/mol tRNA) than for S. cereviseae tRNAAla (0.7 mol I/mol tRNA). Titration experiments yielded an ≈15- to 20-fold difference in enzymatic activities (data not shown). The nature of the modified base was confirmed to be inosine by two-dimensional TLC (data not shown) with added unlabeled IMP as an internal standard. The possibility that hADAT1 might also catalyze the second step of the A37 → m1-I37 conversion in tRNAAla through an intrinsic methyltransferase activity was tested by adding S-adenosylmethionine to the in vitro reaction. No m1-I was detected after two-dimensional TLC analysis of the in vitro-incubated and P1 nuclease-digested tRNAAla. To reconfirm the specific deaminase activity of hADAT1 on the human tRNA transcript (of which the corresponding gene is as yet uncharacterized), we constructed the gene deduced from the mature tRNAAla sequence. Reverse transcription–PCR and sequencing of in vitro-assayed transcripts demonstrated the specific A37 → I37 conversion by hADAT1 (Fig. 4B), whereas the point mutant hADAT1-QAD lacked detectable deaminase activity (data not shown).
To further characterize the determinants for substrate specificity of hADAT1, we constructed a synthetic RNA template of 17 nt representing the anticodon stem–loop region of human tRNAAla and assayed for specific modification of A37 after incubation with hADAT1, ADAR1, ADAR2, and the described chimeras (Fig. 3) in vitro. No deamination of A37 was detected after 2.5 h of incubation and reverse transcription–PCR analysis (data not shown). Finally, a chimeric substrate of the 17-nt anticodon stem–loop of tRNAAla fused to 22 bp of dsRNA was not a substrate for modification at A37 by the ADAR2/hADAT1 fusion constructs (Fig. 3; data not shown).
DISCUSSION
In this report, we describe the cDNA isolation and functional characterization of hADAT1, a human adenosine deaminase specific for the conversion of A37 to I37 in eukaryotic tRNAAla. Initiated by sequence homology searches for potential RNA editing enzymes related to the ADAR family of adenosine deaminases we noticed a human expressed sequence tag (accession no. AA161179) with high sequence similarity to a short stretch of key residues common to the catalytic domain of ADAR proteins. This sequence information was used to isolate the corresponding human cDNA. The analysis of the deduced 502-aa ORF confirmed the presence of a structural domain closely related to the adenosine deaminase catalytic region of ADARs (Fig. 1). However, hADAT1 lacks dsRNA-binding domains present in all other mammalian ADAR proteins characterized to date and harbors a stretch of 90 aa embedded in the catalytic deaminase region that is unrelated to any sequence in the databases. The relative position of this extra domain inside the deaminase region is equivalent to sites in ADAR2 and RED2 where, through alternative splicing, 40 (ADAR2) or 10 (RED2) aa can be inserted without abolishing deaminase function (shown only for ADAR2) (18, 32). However, in the case of hADAT1, no alternative splicing could be detected by PCR analysis on human cDNAs from brain and lung tissues. Furthermore, PCR on human genomic DNA confirmed that this domain insert is not encoded by a separate exon and thus cannot be spliced without also deleting essential parts of the deaminase domain (S.M. and A.R., unpublished results). It may be a protein–protein interaction domain for contacting other tRNA modifying enzymes, possibly the methylase catalyzing the second step of the A37 conversion to m1-I37. Alternatively, it might confer RNA-binding activity on hADAT1, although it appears unlikely because the functional homologue from yeast, Tad1p, lacks such a domain. Rather, the RNA-binding domain of hADAT1 and Tad1p may overlap with the catalytic deaminase region similar as in APOBEC-1 (apolipoprotein B editing enzyme catalytic polypeptide 1), where the cytidine deaminase domain harbors residues that bestow a low-affinity RNA-binding activity (reviewed in ref. 33; see also ref. 34).
Fig. 5 shows an alignment of all known human ADAR proteins as well as related proteins and sequences retrieved from the databases including the characteristic signature motifs common to all RNA-specific adenosine deaminases. With the knowledge of two tRNA specific enzymes, hADAT1 and the yeast protein Tad1p, we are able to delineate sequence motifs that might be indicative of their specific function (Fig. 1 and 5). Clearly, the D. melanogaster ORF deduced from the genomic sequence DS00941 is likely to be the functional homologue of hADAT1 and Tad1p, whereas another D. melanogaster-derived genomic sequence (N35H14) shares sequence motifs common to the pre-mRNA specific subfamily of ADARs. From these alignments it is suggested that the genomic sequences from Trypanosoma brucei and Candida albicans probably represent genes of the respective tRNA specific adenosine deaminases of these species rather than an ADAR protein acting on mRNA (Fig. 5).
Figure 5.
Multiple sequence alignment of known and putative ADAR deaminase domains. The EMBL/GenBank accession numbers are as follows: hADAR1, hADAR2a, rRED2, hADAT1, DS00941 and Tad1p, see Fig. 1; mTenr, accession no. X84693; T20H4.4, U00037; N35H14, AL035207. See Materials and Methods for source of C. albicans (C.a. 2–4410) and T. brucei (T.b. 12A2) sequences. Highly conserved residues (identical or similar between dsRNA specific and tRNA specific ADARs) are highlighted in black and gray. Putative signature motifs of characterized and putative tRNA-specific adenosine deaminases (boxed with dashed line) and conserved motifs indicative for pre-mRNA specific ADARs (boxed) are depicted.
The observed transcript lengths of hADAT1 (5.0 and 6.5 kb) expressed in human tissues are significantly longer than the 1.5-kb ORF and likely reflect the presence of an extended 3′-untranslated region, because no canonical polyadenylation signal was included on the cDNA clones and the 5′-untranslated sequence deduced from all isolated cDNA clones and from 5′-rapid amplification of cDNA ends experiments was shorter than 200 bp.
In vitro, recombinantly expressed and purified hADAT1 did not have detectable adenosine deaminase activity on RNA molecules known to be substrates for modification by ADAR1 or ADAR2. Because the addition of pure hADAT1 to cellular extracts from human embryonic kidney cells did also not induce adenosine deamination in these RNAs above background levels (S.M. and A.R., unpublished data), no cofactors seem to exist that might confer dsRNA-binding activity on hADAT1. These results, together with earlier observations that no inosine was generated by ADAR1 or ADAR2 incubated with tRNA substrates (27), reflects the different requirements for specific recognition of dsRNA structures characterized by A-form double helices with bulges and loops on one hand (ADAR1, ADAR2) and the unique L-shaped three-dimensional structure of tRNA (hADAT1, Tad1p) on the other. Clearly, the anticodon stem–loop of tRNAAla alone is not a functional substrate for hADAT1 in vitro, and chimeric proteins fusing dsRNA-binding domains to hADAT1 do not target A37 within the tRNAAla anticodon stem–loop extended by a region of dsRNA.
We have demonstrated that hADAT1 in vitro specifically and with high efficiency deaminates A37 in tRNAAla from higher (human, B. mori) and with reduced efficiency from lower (S. cereviseae) eukaryotes. This is in accordance with the observations of the yeast Tad1p deaminase being more active on tRNAAla from yeast than tRNAAla from B. mori (27) and can be attributed to the differences in sequence and thus three-dimensional structure of the substrates. This is further substantiated by our observation that hADAT1 does not detectably target A37 in yeast tRNAAla in vivo when expressed in a Tad1-deficient yeast strain even though the enzyme was expressed at high levels and the corresponding yeast extract was highly active on A37 in human tRNAAla assayed in vitro (unpublished data).
The function of tRNA nucleotides proximal to the anticodon is not fully understood (reviewed in ref. 35). They have been implicated as discrimination sites for recognition by tRNA synthetases (36) and may influence the stability of the tRNA binding to its complementary codon in mRNA (37). The m1-I37 modification, unique to eukaryotic tRNAAla, is likely to be of physiological significance because our results demonstrate that the specific machinery for its synthesis has been conserved throughout eukaryotic evolution. It may help to prevent translational frameshifting, as was demonstrated for the modification m1-G37 in tRNAs from the prokaryote Salmonella typhimurium (38). Even though the ablation of the Tad1p gene in S. cereviseae did not result in an obvious phenotype (27), it might be possible to assess the importance of m1-I37 in tRNAAla for maintaining translational accuracy in vivo by using appropriate reporter systems (38) and the TAD1-disrupted yeast strain.
It is not known how many tRNAAla genes exist in humans. A single tRNAAla genomic sequence has been reported to date (39). However, it does not correspond to the tRNAAla transcript sequences characterized (25) and is not a substrate for hADAT1 when transcribed in vitro (S.M. and A.R., unpublished data). It may represent a pseudogene (39). Thus, if all expressed human tRNAAla genes give rise to transcripts that undergo A37 → m1-I37 modification, it will be interesting to see what global or tissue-specific consequences are found with functional inactivation of the ADAT1 gene in a mammalian organism.
Acknowledgments
We thank W. Keller for his support and helpful discussions. We also thank Dr. K. Nishikura for hADAR1-cDNA, Dr. P. H. Seeburg for rADAR2 cDNA, and Dr. H. Grosjean for B. mori and yeast tRNAAla gene constructs and mutants. We are grateful to Dr. B. Johnson for advice with insect cell culture and to Dr. S. Bell for Sf9 cells. We acknowledge a long-term fellowship of the European Molecular Biology Organization to S.M. and a graduate student fellowship from Boehringer Ingelheim Fonds to A.P.G. This work was supported by grants from National Institutes of Health, National Science Foundation, and the National Foundation for Cancer Research.
ABBREVIATIONS
- ADAR
adenosine deaminase acting on RNA
- hADAT1
human adenosine deaminase acting on tRNA
- dsRNA
double-stranded RNA
- kb
kilobase
- A
adenosine
- I
inosine
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF125188).
References
- 1.Bass B L. Trends Biochem Sci. 1997;22:157–162. doi: 10.1016/s0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- 2.Maas S, Melcher T, Seeburg P H. Curr Opin Cell Biol. 1997;9:343–349. doi: 10.1016/s0955-0674(97)80006-3. [DOI] [PubMed] [Google Scholar]
- 3.Rueter S M, Emeson R B. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 343–361. [Google Scholar]
- 4.Maas S, Melcher T, Herb A, Seeburg P H, Keller W, Krause S, Higuchi M, O’Connell M A. J Biol Chem. 1996;271:12221–6. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 5.Basilio C, Wahba A J, Lengyel P, Speyer J F, Ochoa S. Proc Natl Acad Sci USA. 1962;48:613–616. doi: 10.1073/pnas.48.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Nature (London) 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 7.Seeburg P H, Higuchi M, Sprengel R. Brain Res Brain Res Rev. 1998;26:217–229. doi: 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- 8.Wagner R W, Nishikura K. Mol Cell Biol. 1988;8:770–777. doi: 10.1128/mcb.8.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melcher T, Maas S, Herb A, Sprengel R, Seeburg P H, Higuchi M. Nature (London) 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 10.Paul M S, Bass B L. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melcher T, Maas S, Higuchi M, Keller W, Seeburg P H. J Biol Chem. 1995;270:8566–8570. doi: 10.1074/jbc.270.15.8566. [DOI] [PubMed] [Google Scholar]
- 12.Rueter S M, Burns C M, Coode S A, Mookherjee P, Emeson R B. Science. 1995;267:1491–1494. doi: 10.1126/science.7878468. [DOI] [PubMed] [Google Scholar]
- 13.Yang J H, Sklar P, Axel R, Maniatis T. Nature (London) 1995;374:77–81. doi: 10.1038/374077a0. [DOI] [PubMed] [Google Scholar]
- 14.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Proc Natl Acad Sci USA. 1994;91:11457–61. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Connell M A, Krause S, Higuchi M, Hsuan J J, Totty N F, Jenny A, Keller W. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hough R F, Bass B L. RNA. 1997;3:356–370. [PMC free article] [PubMed] [Google Scholar]
- 17.Mittaz L, Scott H S, Rossier C, Seeburg P H, Higuchi M, Antonarakis S E. Genomics. 1997;41:210–217. doi: 10.1006/geno.1997.4655. [DOI] [PubMed] [Google Scholar]
- 18.Gerber A, O’Connell M A, Keller W. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 19.Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg P H. J Biol Chem. 1996;271:31795–8. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 20.Higuchi M, Single F N, Kohler M, Sommer B, Sprengel R, Seeburg P H. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 21.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger J R, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg P H. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 22.Herb A, Higuchi M, Sprengel R, Seeburg P H. Proc Natl Acad Sci USA. 1996;93:1875–1880. doi: 10.1073/pnas.93.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holley R W. J Am Med Assoc. 1965;194:868–871. [PubMed] [Google Scholar]
- 24.Grosjean H, Auxilien S, Constantinesco F, Simon C, Corda Y, Becker H F, Foiret D, Morin A, Jin Y X, Fournier M, Fourrey J L. Biochimie. 1996;78:488–501. doi: 10.1016/0300-9084(96)84755-9. [DOI] [PubMed] [Google Scholar]
- 25.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auxilien S, Crain P F, Trewyn R W, Grosjean H. J Mol Biol. 1996;262:437–458. doi: 10.1006/jmbi.1996.0527. [DOI] [PubMed] [Google Scholar]
- 27.Gerber A, Grosjean H, Melcher T, Keller W. EMBO J. 1998;17:4780–4789. doi: 10.1093/emboj/17.16.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 30.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 31.Sorrentino B P, McDonagh K T, Woods D, Orlic D. Blood. 1995;86:491–501. [PubMed] [Google Scholar]
- 32.Lai F, Chen C X, Carter K C, Nishikura K. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith H C, Gott J M, Hanson M R. RNA. 1997;3:1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 34.Navaratnam N, Fujino T, Bayliss J, Jarmuz A, How A, Richardson N, Somasekaram A, Bhattacharya S, Carter C, Scott J. J Mol Biol. 1998;275:695–714. doi: 10.1006/jmbi.1997.1506. [DOI] [PubMed] [Google Scholar]
- 35.Bjork G R. In: tRNA: Structure, Biosynthesis and Function. Soll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 165–205. [Google Scholar]
- 36.Rould M A, Perona J J, Steitz T A. Nature (London) 1991;352:213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- 37.Labuda D, Striker G, Grosjean H, Porschke D. Nucleic Acids Res. 1985;13:3667–3683. doi: 10.1093/nar/13.10.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bjork G R, Wikstrom P M, Bystrom A S. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 39.Buckland R A, Maule J C, Sealey P G. Genomics. 1996;35:164–171. doi: 10.1006/geno.1996.0335. [DOI] [PubMed] [Google Scholar]