Abstract
Background: Cytogenetic studies of malignant peripheral nerve sheath tumours (MPNSTs) and malignant triton tumours (MTTs) are rare.
Aims: To undertake cytogenetic analysis of these tumours.
Methods: Conventional cytogenetic analysis of 21 MPNSTs and MTTs from 17 patients (nine with peripheral neurofibromatosis (NF1)) was carried out using standard culture and harvesting procedures. For a more precise identification of composite structural rearrangements and marker chromosomes, spectral karyotypic analysis (SKY) was applied to a subset of cases. In addition, EGFR gene copy number was assessed by fluorescence in situ hybridisation (FISH) analysis in a subset of cases.
Results: Cytogenetic analysis revealed predominantly complex karyotypes. SKY analysis was useful in further defining many structural anomalies. Structural aberrations most frequently involved chromosomal bands or regions 1p31–36, 4q28–35, 7p22, 11q22–23, 19q13, 20q13, and 22q11–13. Overall, loss of chromosomal material was much more common than gain. Loss of chromosomes or chromosomal regions 1p36 (48%), 3p21–pter (52%), 9p23–pter (57%), 10 (48%), 11q23–qter (48%), 16/16q24 (62%), 17(43%), and 22/22q (48%), and gains of 7/7q (29%) and 8/8q (29%) were most prominent. These gains and losses were distributed equally between MPNST and MTT, demonstrating that these entities are similar with respect to recurrent genomic imbalances. Similarly, none of the recurrent chromosomal breakpoints or imbalances was restricted to either NF1 associated or sporadic MPNSTs. FISH analysis was negative for amplification.
Conclusions: These cytogenetic and molecular cytogenetic findings expand the knowledge of chromosomal alterations in MPNST and MTT, and point to possible recurring regions of interest.
Keywords: cytogenetic, malignant peripheral nerve sheath tumour
Malignant peripheral nerve sheath tumours (MPNSTs) are tumours that arise from Schwann cells, frequently in association with peripheral nerves or in an existing neurofibroma. MPNSTs comprise 5–10% of all soft tissue sarcomas.1 Malignant triton tumour (MTT) is a rare histological variant of MPNST characterised by rhabdomyoblastic differentiation.2 Most MPNSTs and MTTs occur in patients with neurofibromatosis 1 (NF1), and both entities are typically associated with a poor prognosis.3
“Few cases of malignant peripheral nerve sheath tumour and malignant triton tumour have been subjected to cytogenetic analysis”
Tumour specific chromosomal abnormalities have been shown to characterise certain subgroups of mesenchymal tumours.4 Unfortunately, few cases of MPNST and MTT have been subjected to cytogenetic analysis and most of those examined have exhibited complex karyotypes, often marked by intratumoral and intertumoral heterogeneity.5,6,7,8,9,10,11 Moreover, spectral karyotypic (SKY) and multifluorochrome in situ hybridisation (M-FISH), molecular cytogenetic approaches that could be useful in deciphering many of the complex structural rearrangements and marker chromosomes featured in MPNST/MTT complements, have been performed on only two cases.9,10 In our current study, 21 MPNST and MTT specimens were examined by SKY, FISH, and/or conventional cytogenetic analysis.
MATERIALS AND METHODS
Tumour samples
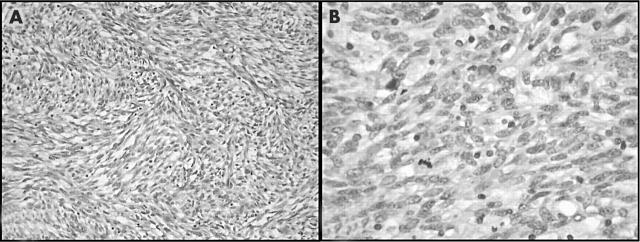
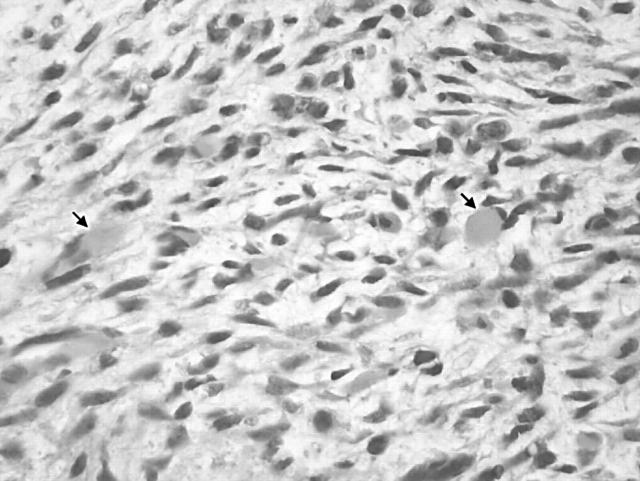
During a 16 year period, representative tissue samples of 14 MPNST and seven MTT specimens from 17 patients (nine with NF1) were received directly or within 36 hours of surgery for cytogenetic analysis. Table 1 lists the clinicohistopathological features of the patients and corresponding tumours; nine patients were female and eight were male, with ages ranging from 2 to 78 years (mean, 35). The mean age of the patients with NF1 was 28 years, compared with 41 for the sporadic cases. Most (14 of 17) of the tumours were located in the extremities. All pathology reports and/or slides were reviewed for accuracy to confirm that the clinical, histological, and immunohistochemical findings supported a diagnosis of MPNST or MTT. Most lesions (cases 1, 3, 4–6, 8, 10–12, 14, 15, and 17) exhibited histological and immunohistochemical features of conventional high grade MPNST using the diagnostic guidelines published in fascicle 24 of the third AFIP series.12 The tumours were composed of enlarged hyperchromatic spindle shaped cells arranged in a fascicular pattern. Mitotic activity was prominent in all cases and geographical necrosis and frank anaplasia were seen in some (fig 1). Patchy immunohistochemical staining for the S-100 protein was present in most cases. The MTTs (cases 7, 9, 13, and 16) were characterised by the presence of rhabdomyoblasts, as demonstrated by smooth muscle actin or desmin immunohistochemistry (fig 2). The rhabdomyoblasts were distinguished from entrapped skeletal muscle. One case (case 2) showed predominantly epithelioid cell morphology (epithelioid MPNST).
Table 1.
Clinicohistopathological and karyotypic data
| Case | Age/Sex | Indication | Location (Size (cm)) | P/R/M | RT-PCR | Karyotype |
| 1a | 45/M | MPNST | Ankle | P (biopsy) | NP | 42,XY,-3,add(4)(q35),add(11)(p14),-12,-14,add(19)(q13.4),-21,-21,-22,-22,+mar1,+mar2,+mar3[9]/42,XY,idem,add(1)(p36),del(7)(q31)[3]/46,XY[11] |
| 1b | 45/M | MPNST | Ankle (6.2×6.0×2.8) | R (resection) | NP | 42,XY,del(1)(q35),del(2)(q23),add(4)(q35),+5,del(7)(q31),-9,-9,add(11)(q23),-12,-19,add(19)(q13.4),-21,-22,+mar1,+mar[cp14]/46,XY[7] |
| 2 | 78/M | Epithelioid MPNST | Thigh/buttock (1.5×1.0×0.5) | P (biopsy) | Negative | 43,XY,i(1)(q10),+8,-9,-11,del(12)(p11.1),-18,-22[6]/46,XY[14] |
| 3* | 37/M | MPNST | Lung (1.8×1.2×1.0) | M (resection) | NP | 65,XXY,add(3)(p11.2),add(5)(q35)×2,del(7)(q11.1q34)×2,add(9)(p23),-10,add(13)(p11.1),add(14)(q32)×2,-16,-19,-19,add(20)(q13.3),add(22)(q11.2)×2,1∼3dmin[cp3]/92,XXYY[4]/46,XY[14] |
| 4* | 16/M | MPNST | Thigh (3.0×2.0×0.5) | P (biopsy) | Negative | 46,XY,t(1;3)(p36.1;q11.2),del(7)(q32),+i(8)(q10),del(9)(p13),-10,del(11)(q14),der(12)t(1;12)(q23;p12),der(14)t(10;14)(q11.2;p11.2), der(15)t(15;21) (q26;q?)?dup(21),add(20)(q13.2),-21,add(22)(q11.2),+mar1, 1∼3dmin[cp17]/92, idem×2[cp3]/46,XY[8] |
| 5 | 72/F | MPNST | Thigh (17×6.5×6.5) | P (resection) | Negative | 72,XXX,+1,add(1)(p31)×2,del(2)(p13p23),del(4)(p14p16)×2,del(6)(q13q26)×2,+7,add(7)(q36)×2,+8,-9,-10,del(11)(q23),+12,-14,-15,-16,-17,-18, +add(19)(q13.3),+20,+21,+mar1,+mar2,+mar3,1∼5dmin[cp8]/46,XX[2] |
| 6 | 28/M | MPNST | Neck (8.0×6.5×2.8) | R (resection) | NP | 60∼70,XY,-X,-1,add(1)(p36.3),-3,-6,-8,-9,-10,add(11)(q22),del(11)(q21),-13,-14,-15,add(15)(q24),add(16)(q21),add(16)(q24),-17,-17,-17,+add(19)(q13.2),-20,+21,add(22)(q11.2),+mar1,+mar2,+mar3,+mar4,+mar5,+mar6×2,+mar7,+mar8,+1∼4mar[cp17] |
| 7 | 28/F | MTT | Axilla (11.0×4.0×3.0) | P (resection) | Negative | 64,XX,-X,+1,der(1)t(1;15)(p11;q11.1)×2,+2,-3,-4,+5,-6,+7,+8,-9,-10,-11,-12,-13,-14,-15,-15,-15,-16,+17,+18,+20,+21[5]/46,XX[15] |
| 8* | 21 months/F | MPNST | Shoulder (2.5×1.5×1.5) | P (biopsy) | Negative | 58,XX,-X,-1,-3,-4,-5,-6,+7,+8,-9,-10,-11,-13,-14,+15,-16,-17,-18,-22,+mar[10] |
| 9a† | 25/F | MTT | Forearm (1.5×1.5×3.0) | R (resection) | NP | 47,XX,t(1;2)(p13;p25),t(3;14)(p12;q31),ins(4;?5)(q28;?q22q32),-5,add(9)(p24),add(15)(q26),add(22)(q13),+mar1,+mar2[5]/47,XX,add(6)(p25),t(7;8)(p11;p11),t(12;21)(q21;q21),+22[4]/47,XX,t(2;10)(q31;p15),t[3;der(7)][p13;q36],der(7)t(7;?7)(p22;?q?),+mar[2]/46,XX[2] |
| 9b | 27/F | MTT | Forearm | R (biopsy) | NP | 44,XX,-2,-3,del(3)(p13),-4,add(6)(q27),der(7)t(7;?7)(p22;?q?),-10,der(12)t(12;21)(q24;q21),add(13)(q34),add(15)(q26),-16,-17,-21,+mar1,+mar2,+mar3,+mar4,+mar5[cp7] |
| 9c | 27/F | MTT | Forearm (5.0×2.0×2.0) | R (resection) | neg. | 45,XX,-2,-3,del(3)(p13),ins(4;?5)(q28;?q22q32),add(6)(q27),der(7)t(7;?7)(p22;?q?),add(13)(q34),add(15)(q26),-16,-17,-21,+mar1,+mar2,+mar3,+mar4[cp19] |
| 10 | 50/F | MPNST | Shoulder (9.0×7.5×7.5) | P (resection) | NP | 71∼82,XXX,del(1)(q21)×2,add(2)(q?),del(4)(p13),-5,-5,+del(7)(p11),-8,-8,-9,-9,-10,-10,del(12)(p11),+14,add(16)(q24),add(20)(q11),+mar1,+mar2,inc[cp3]/46,XX[9] |
| 11* | 26/M | MPNST | Pelvis (19.0×10.0×7.5) | P (resection) | Negative | 71-75,XY,-X or -Y,-3,dic(3;9)(q11;p11)×2,-4,del(4)(q28q35),der(7)del(7)(q22)t(1;17;7)(?;?;p22)×2,der(7)t(7;7)(p22;?),+der(7)t(7;7)(p22;?),-8,add(8)(q24.3)×2,-9,-9,+11,del(11)(q23)×2,del(12)(q24), +der(14)t(7;14)(?;p11),-15,-16,-16,-16,-17,der(17)del(17)(p13)t(17;17)(q25;?)×2,+18,der(19)t(13;19;21)(q?;?p11q11;q?)×2,der(20)t(12;20)(?q13;p13)×2,der(21)t(17;21)(?;q22)×2,der(22)t(8;22)(?;p11),der(22)t(8;17;22)(?;?;p11), +mar1,+mar2,+mar3×2,+5∼9mar[cp20]/46,XY[2] |
| 12*† | 35/F | MPNST | Foot (4.5×3.0×3.0) | P (biopsy) | Negative | 47∼65,XX,del(1)(p31),+del(3)(q13.3),+del(6)(q21)×2,+del(6)(q27)×2,del(7)(q21),add(12)(q24),+add(16)(q24),add(20)(q11.2),-21,+mar1,+mar2,+6mar[cp3]/46,XX[44] |
| 13a*† | 17/F | MTT | Knee | P (biopsy) | Negative | 69,XX,-X,del(1)(p21),-3,+der(5)t(5;?9)(p13;?q13),der(5)t(5;?9)(p13;?q13),+7,+8,+8,-9,-9,-9,-10,-10,dup(11)(p11.2p15),+del(12)(q15),del(12)(q15),-13,-14,-16,-17,-18,+21,-22,+mar1×2,+mar2,+mar3,+3mar[cp20]/46,XX[12] |
| 13b*† | 17/F | MTT | Knee (5.5×5.5×20.0) | P (resection) | Negative | 64,XX,?del(X)(q22),+?del(X)(q22),-1,-2,der(5)t(5;?9)(p13;?q13),+7,-9,-10,-11,-12,-16,-17,-18,+mar1,+mar1[12]/42,X,del(X)(q22),-1,-3,der(5)t(5;?9)(p13;?q13),-9,-11,-14,-17,-18,+mar1,+mar2,+mar3[7]/46,XX[1] |
| 14* | 60/M | MPNST | Lumbar region (25.0×23.0×10.0) | P (resection) | NP | 44,XY,del(3)(p21),del(10)(q22),-13,-16,del(20)(q13.3),add(22)(q11.2)[3]/46,XY[21] |
| 15* | 32/F | MPNST | Upper arm (4.5×2.5×2.0) | P (resection) | Negative | 46,XX,add(1)(q21),-6,-10,-14,-18,+mar1,+mar2,+mar3,+mar4,inc[cp12]/46,XX[9] |
| 16† | 27/F | MTT | Paraspinal (5.6×8.4×14.3) | P (resection) | NP | 63,XX,-X,+dic(1;22)(p36;p10),+del(2)(q33),t(2;?12)(p25;?),-3,add(4)(q34),+5,dic(7;?11)(p22;?),-10,-11,der(11)t(5;11)(q11;q23),-12,-12,-14,add(14)(p11),-15,-16,add(16)(p13),-17,-18,-19,-20,del(20)(q13),?add(21)(q22),+mar1,+mar2,+mar3,+mar4,2∼8dmin[cp5] |
| 17* | 20/M | MPNST | Lower leg (28×20×15 cm) | P (resection) | NP | 67,XXY,+add(1)(p35),+3,-13,add(19)(q13),-20,-20,-21[8]/46,XY[13] |
Figure 1.
Case 5: malignant peripheral nerve sheath tumour. (A) A low power view shows a cellular spindle cell neoplasm arranged in fascicles (haematoxylin and eosin (H&E) staining). (B) Under higher power, mitotic activity and cytological atypia can be seen (H&E staining).
Figure 2.
Case 7: malignant triton tumour. Spindle cell sarcoma with rhabdomyoblastic differentiation (arrows) (haematoxylin and eosin staining).
Cytogenetic analysis
Cytogenetic analysis was performed by means of standard culture and harvesting procedures, as described previously.6 Briefly, sterile representative tumour tissue was disaggregated mechanically and enzymatically, and then cultured in RPMI 1640 medium supplemented with 20% fetal bovine serum and 1% penicillin/streptomycin-L-glutamine (Irvine Scientific, Santa Ana, California, USA) for three to five days. Two to four hours before harvesting, the cells were exposed to Colcemid (0.02 μg/ml). After hypotonic treatment (0.074 M KCl for 30 minutes for flasks and 0.8% sodium citrate for 25 minutes for coverslips), the preparations were fixed three times with methanol/glacial acetic acid (3/1). Metaphase cells were banded with Giemsa trypsin, and the karyotypes were expressed in accordance with the International System for Human Cytogenetic Nomenclature 1995.13
Molecular cytogenetic analysis
To determine the presence or absence of EGFR (epidermal growth factor receptor) gene amplification, bicolour FISH studies using the LSI EGFR/CEP 7 dual colour probe (Vysis, Downers Grove, Illinois, USA) were performed on cytological touch preparations of four MPNST/MTT specimens (cases 4, 5, 12, and 13a) that exhibited double minutes or marker chromosomes cytogenetically and had sufficient tissue available for analysis. This probe set is composed of a Spectrum Green labelled chromosome 7 specific centromeric probe and a Spectrum Orange labelled EGFR locus specific identifier DNA probe. As negative controls, cytological touch preparations of normal skeletal muscle and peripheral blood lymphocytes were simultaneously hybridised with these probes. All slides were stored at −20°C before analysis. After pretreatment of the slides, the cells and probe were codenatured for one minute at 75°C and incubated at 37°C overnight using HYBrite™ (Vysis). Images were prepared using the Cytovysion image analysis system (Applied Spectral Imaging, Pittsburgh, Pennsylvania, USA). The numbers of hybridisation signals for each specimen were assessed in 200 interphase nuclei by two different individuals. The chromosome 7 centromeric signal was used to determine chromosome 7 copy number.
Spectral karyotyping was performed on unstained cytogenetic preparations of cases 4, 6, and 11, according to the manufacturer’s recommended protocol (Applied Spectral Imaging, Carlsbad, California, USA), and as described previously.14 Spectral images were acquired and analysed with an SD 200 spectral bioimaging system (Applied Spectral Imaging) attached to a Zeiss microscope (Axioskop 2). The spectral image was generated by acquiring approximately 100 frames of the same image that differ only in the optical path difference. Image acquisition was achieved using the Spectral Imaging software (version 2.3; Applied Spectral Imaging). The images were stored for further analysis using the Sky View software (version 1.6.2; Applied Spectral Imaging). Five metaphase cells were analysed for each specimen.
Reverse transcriptase polymerase chain reaction
Reverse transcriptase polymerase chain reaction (RT-PCR) studies were performed on all cases with available tissue to exclude the possibility of a cryptic t(X;18)(p11.2;q11.2) associated fusion transcript (SYT–SSX1 and SYT–SSX2). Total RNA was extracted from representative snap frozen or paraffin wax embedded tissue from seven MPNST (cases 2, 4, 5, 8, 11, 12, and 15) and four MTT (cases 7, 9, 13a, and 13b) specimens with Trizol reagent (Gibco BRL, Gaithersburg, Maryland, USA). Total RNA (1 μg) was reverse transcribed with random hexamers using 200 units of Superscript II RT (Gibco BRL). The resulting cDNAs were PCR amplified using SYT–SSX primers, as described previously.15 Forty thermal cycles were used at the following temperatures: denaturation at 94°C for one minute, annealing at 62°C for one minute, and elongation at 72°C for one minute. The amplified fragments were identified by gel electrophoresis and ethidium bromide staining. The integrity of the mRNA was assessed by an independent amplification using primers to the ubiquitously expressed hypoxanthine phosphoribosyl transferase gene.15 The positive controls were known SYT–SSX1 and SYT–SSX2 fusion transcript positive synovial sarcomas (also cytogenetically t(X;18)(p11.2;q11.2) confirmed). A negative control devoid of a template and a negative control lacking reverse transcriptase were included in all RT-PCR studies.
RESULTS
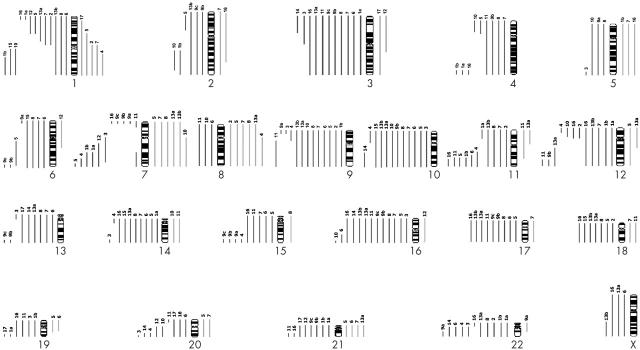
Conventional cytogenetic analysis demonstrated complex chromosomal complements in all 21 specimens (table 1). The cytogenetic abnormalities seen in MPNST and MTT were similar. The modal chromosome number varied, with near triploid complements in 12 specimens and near diploid in nine. Unrelated clones or subclones were found in four specimens. Figure 3 illustrates the chromosomal gains and losses for each individual specimen. Chromosomal loss was more frequent than gain. In particular, loss of whole (or most of) chromosomes or chromosomal regions 1p36 (10 of 21 specimens), 3p21–pter (11 of 21 specimens), 9p23–pter (12 of 21 specimens), 10 (10 of 21 specimens), 11q23–qter (10 of 21 specimens), 16/16q24 (13 of 21 specimens), 17 (nine of 21 specimens), and 22/22q (10 of 21 specimens), and gains at 7/7q and 8/8q (six of 21 specimens each) were most prominent.
Figure 3.
Schematic illustration of the chromosomal gains (bars to the right of each chromosome) and chromosomal losses (bars to the left of each chromosome) detected in each specimen.
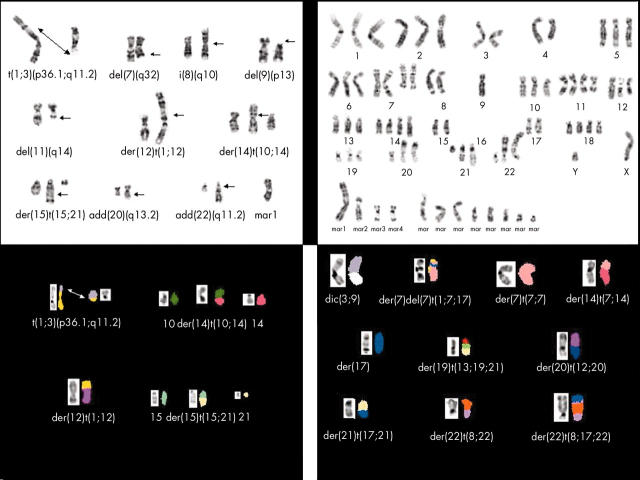
The structural rearrangements among the 21 specimens resulted in 85 breakpoints that could be assigned to specific chromosomal bands. The breakpoints involved every chromosome with the exception of the Y chromosome and chromosome 18. The chromosomal bands/regions most frequently involved in structural rearrangements were: 1p31–36 (seven cases); 7p22, 11q22–23, and 22q11–13 (five cases each); 4q28–35, 19q13, and 20q13 (four cases each); and 6q26–27, 12p11–12, 12q24, 14p11, 16q24, and 21q21–22 (three cases each). Marker chromosomes were observed in most specimens (16 of 21). Double minutes, cytogenetic manifestations of genomic amplification, were detected in four of 21 specimens. FISH analysis of cases 4, 5, 12, and 13a for EGFR gene copy number failed to reveal gene amplification, although 1–2 extra copies of the EGFR gene locus were seen in 25% of the cells examined for case 4 (EGFR to CEP7 ratio, < 2). SKY analyses of cases 4 and 11 were useful in characterising anomalies not identifiable by conventional cytogenetic analysis. For example, without the assistance of SKY analysis, the complex translocation involving chromosomes 15 and 21 [der(15)t(15;21)(q26;q21)dup(21)(q21q11.2)] in case 4 and the complex translocation in case 11 involving chromosomes 1, 7, and 17 [der(7)del(7)(q22)t(1;17;7)(?;?;p22)], originally assigned as marker chromosomes, could not have been defined (fig 4). Unfortunately, however, SKY analysis was unsuccessful in case 6, despite repeated efforts. RT-PCR analysis was negative for the presence of the SYT–SSX1 and SYT–SSX2 fusion transcripts in all specimens evaluated (table 1).
Figure 4.
Representative Giemsa trypsin banding and spectral karyotypic images of cases 4 (upper and lower left) and 11 (upper and lower right), illustrating select complex structural rearrangements and marker chromosomes delineated by spectral karyotypic analysis analysis.
DISCUSSION
MPNSTs most often arise from neurofibromas, often of the plexiform type.3 MTT is a rare MPNST histological variant that shows rhabdomyosarcomatous differentiation.2 MPNST and MTT commonly arise in patients with the autosomal dominant disorder NF1, but can also occur sporadically. Various germline mutations of the NF1 gene, localised to 17q11.2 and encoding the RasGTPase activating protein neurofibromin, have been demonstrated in families affected by NF1.3,16,17 Loss of heterozygosity or the presence of a mutation of the second allele of NF1 has been demonstrated in MPNSTs, lending support to the premise that NF1 acts as a tumour suppressor gene. The risk of local recurrence and metastasis for patients with either NF1 associated or sporadically occurring MPNSTs and MTTs is high, and the overall longterm survival is poor.1,3,18
Cytogenetic analysis has contributed greatly to our understanding of the biological events underlying the origin and progression of many mesenchymal tumours and has identified tumour specific structural rearrangements, particularly chromosomal translocations, that may serve as aids in the diagnosis and classification of these tumours.19 Unfortunately, cytogenetic studies of MPNST and MTT are few.5,6,7,8,9,10,11 In our study, conventional cytogenetic analysis of 21 MPNST and MTT specimens revealed karyotypes featuring multiple numerical and structural abnormalities, a finding similar to previous reports.
Provision of a complete karyotype for MPNST and MTT has been frequently hampered by suboptimal morphology and the complexity of the aberrations present. Molecular cytogenetic approaches such as FISH and SKY are useful adjuncts in further defining abnormalities in these circumstances. In particular, SKY enables the unambiguous identification of metaphase chromosomes by displaying each chromosome in a unique colour. To date, only two cases of MPNST/MTT have been subjected to SKY analysis.9,10 Because of the high degree of structural complexity seen in the chromosomal complements of our present report, spectral karyotypic analysis was performed when possible to provide a more precise chromosomal identification of many of the composite structural rearrangements and marker chromosomes. In addition, by defining equivocal regions involved in structural aberrations, SKY analysis allowed more accurate assessment of how the observed structural alterations lead to genomic imbalances. For example, SKY analysis was useful in characterising a t(1;3)(p36.1;q11.2) translocation previously recognised as add(1)(p36) and add(3)(q12), a del(11)(q14.1) formerly classified as a marker chromosome, and a complex translocation involving chromosomes 15 and 21 [der(15)t(15;21)(q26;q21)dup(21)(q21q11.2)] in case 4.
“By defining equivocal regions involved in structural aberrations, spectral karyotypic analysis allowed more accurate assessment of how the observed structural alterations lead to genomic imbalances”
Overall, we found that the following chromosomal breakpoints were most commonly involved in structural rearrangements: 1p31–36, 4q28–35, 7p22, 11q22–23, 19q13, 20q13, and 22q11–13. Plaat et al performed a computer assisted cytogenetic review of 47 MPNST and four MTT cases and concluded that 1p13, 1q21, 7p22, 9p11, 17p11, 17q11, and 22q11 were the most commonly involved breakpoints.7 Subsequently, in a series of 20 MPNSTs, Mertens et al reported that chromosomal breakpoints at 1p11, 5p15, 5q13, 6q21, 8q10, 11q13, 11q21, 14q24, 17q21, 17q25, and 20q13 were most frequently aberrant.8 Clearly, a pattern of recurrent breakpoints appears to be emerging in MPNST, with the involvement of certain chromosomal bands/regions, particularly 1p, 7p22, 11q13–23, 20q13, and 22q11–13, being most prominent.
Interestingly, 7p22 is the locus for ETV1, a member of the ETS family of genes involved in another type of sarcoma (the 7;22 variant translocation detected in a subset of Ewing’s sarcomas20) and the mismatch repair gene PMS2.21 Germline mutations of PMS2 have been detected in some families with Turcot’s syndrome featuring glioblastoma multiforme, colorectal tumours, and café au lait spots.22 Interestingly, the protooncogene SRC (human homologue of the v-src gene of the Rous sarcoma virus) is localised to 20q12–13.23 Recently, activating mutations of the SRC gene have been shown to play a role in the malignant progression of human colon cancer.24
In 1993, Rouleau et al established that the NF2 gene, localised to 22q12.2, is the site of the mutations causing neurofibromatosis 2 (NF2) by demonstrating germline and somatic mutations in patients with NF2 and in NF2 related tumours.25 The role, if any, that NF2 plays in the pathogenesis and/or progression of MPNST has not been established. Some have suggested that loss of 22q11–13 is associated with the development of MPNST,26 whereas recent FISH studies have shown 22q polysomies or gains to be more common in MPNST.27 As mentioned, in our current study and in previous series, structural rearrangements of 22q11–13 were prominent in addition to loss of 22q.7 The apparent discrepancy between conventional cytogenetic and interphase cytogenetic studies may be explained by the presence of unidentifiable marker chromosomes in some karyotypes.28 Additional molecular studies are needed to clarify whether NF2 contributes to the origin or evolution of MPNST.
A review of the genomic imbalances in our series and those previously reported confirms that loss of chromosomal material is far more frequent than gain in MPNST and MTT.5,6,7,8,9,10,11 In particular, loss of whole (or large sections of) chromosomes or chromosomal regions 1p36, 9p, 10, 11q21–24, 16/16q24, 17, and 22/22q, and gains of 7/7q and 8/8q appear to be most prominent. The observation of recurrent 9p loss is compatible with molecular studies showing homozygous deletions of the CDKN2A gene (9p21) encoding the p16 cell cycle inhibitory molecule in MPNSTs.27–29 Similarly, chromosome 17 loss parallels molecular evidence of allelic loss at the NF1 locus (17q11.2) in sporadic and NF1 associated MPNSTs, in addition to functional inactivation of the TP53 tumour suppressor gene localised to 17p13.30–33 The NF1 gene encodes neurofibromin, a multidomain molecule with the capacity to regulate several intracellular processes, including the extracellular signal regulated kinase (ERK) cascade.34 The ERK gene is located at 1p36.1,35 a locus commonly lost in MPNST/MTT. Dysfunction of this signal transduction cascade has been shown in cognitive defects, including mental retardation caused by mutation of the NF1 gene.34 Monosomy 10 has been described in MPNST, and was found in our present study. The presence of the tumour suppressor gene PTEN at 10q23.3 suggests a possible role in MPNST, although one recent study failed to demonstrate a mutation in the PTEN gene in MPNST.11 Lastly, gain of chromosome 7 corresponds to FISH studies showing polysomy 7 and low level EGFR amplification.27 Notably, in contrast to the study by Plaat et al,7 there were no detectable differences in affected breakpoints or chromosomal imbalances between NF1 associated MPNSTs and MTTs and sporadic MPNSTs and MTTs. Moreover, EGFR gene amplification was not detected by FISH in the cases examined here, although one case did exhibit gain of one to two extra copies of the EGFR gene locus.
The differential diagnosis of MPNST frequently includes synovial sarcoma. The presence of an X;18 translocation cytogenetically or an SYT–SSX fusion transcript molecularly has been described in a few cases of MPNST.36 In our current study, RT-PCR analysis of 12 cases in which adequate material was available failed to reveal either an SYT–SSX1 or SYT–SSX2 fusion transcript in the cases analysed. Moreover, none of the MPNST or MTT cases harboured the chromosomal translocation t(X;18)(p11.2;q11.2).
In conclusion, a completely accurate assessment of the frequency of various aberrations in MPNST and MTT is limited by the high percentage of marker chromosomes (chromosomes of unknown origin). A combined approach that includes conventional cytogenetics and molecular cytogenetics such as spectral karyotypic analysis increases the sensitivity of recurrent breakpoint and aneuploidy detection in MPNST. The cytogenetic and molecular cytogenetic findings of the current study and a review of the literature indicate that although MPNST and MTT exhibit complex chromosomal complements, anomalies of certain chromosomal breakpoints and/or the presence of certain chromosomal imbalances appear to be recurrent and, thus, potentially play a role in the pathogenesis or progression of MPNST and MTT.
Take home messages.
We used a combination of conventional cytogenetic and molecular cytogenetic analysis to detect recurrent breakpoints and aneuploidy in malignant peripheral nerve sheath tumour (MPNST) and malignant triton tumour (MTT)
We found that although MPNST and MTT exhibit complex chromosomal complements, anomalies of certain chromosomal breakpoints and/or the presence of certain chromosomal imbalances were recurrent and probably play a role in the pathogenesis or progression of these lesions
Chromosomal bands/regions 1p, 7p22, 11q13–23, 20q13, and 22q11–13 were the most frequently involved breakpoints in MPNST
Loss of chromosomal material was more frequent than gain in MPNST and MTT
Abbreviations
EGFR, epidermal growth factor receptor
ERK, extracellular signal regulated kinase
FISH, fluorescent in situ hybridisation
MPNST, malignant peripheral nerve sheath tumour
MTT, malignant triton tumour
NF, neurofibromatosis
RT-PCR, reverse transcription polymerase chain reaction
SKY, spectral karyotypic analysis
The authors would like to thank J Bailey, P Cattano, and M Nelson for their expert technical assistance and K Christian for her expert secretarial assistance. This work was supported in part by the following: NIH/NCI (P30 CA 36727), JA Wiebe Jr Children’s Health Care Fund, and State of Nebraska, LB595 (JAB).
REFERENCES
- 1.King AA, DeBaun MR, Riccardi VM, et al. Malignant peripheral nerve sheath tumors in neurofibromatosis 1. Am J Med Genet 2000;93:388–92. [PubMed] [Google Scholar]
- 2.Woodruff JM, Chernick NL, Smith MC, et al. Peripheral nerve tumors with rhabdomyosarcomatous differentiation (malignant “triton” tumors). Cancer 1973;32:426–39. [DOI] [PubMed] [Google Scholar]
- 3.Woodruff JM, Kourea HP, Louis DN, et al. Malignant peripheral nerve sheath tumor (MPNST). In: Kleihues P, Cavenee WK, eds. WHO classification of tumours: pathology and genetics. Tumours of the nervous system. Lyon, France: IARC Press, 2000:172–3.
- 4.Bridge JA. Soft tissue sarcomas. In: Wolman SR, Sells S, eds. Cytogenetic markers of human disease. Clifton, NJ: Humana Press, 1997:425–59.
- 5.Travis JA, Sandberg AA, Neff JR, et al. Cytogenetic findings in malignant triton tumor. Genes Chromosomes Cancer 1994;9:1–7. [DOI] [PubMed] [Google Scholar]
- 6.McComb EN, McComb RD, DeBoer JM, et al. Cytogenetic analysis of a malignant triton tumor and a malignant peripheral nerve sheath tumor and a review of the literature. Cancer Genet Cytogenet 1996;91:8–12. [DOI] [PubMed] [Google Scholar]
- 7.Plaat BE, Molenaar WM, Mastik MF, et al. Computer-assisted cytogenetic analysis of 51 malignant peripheral-nerve-sheath tumors: sporadic vs. neurofibromatosis-type-1-associated malignant schwannomas. Int J Cancer 1999;83:171–8. [DOI] [PubMed] [Google Scholar]
- 8.Mertens F , Dal Cin P, De Wever I, et al. Cytogenetic characterization of peripheral nerve sheath tumours: a report of the CHAMP study group. J Pathol 2000;190:31–8. [DOI] [PubMed] [Google Scholar]
- 9.Haddadin MH, Hawkins AL, Long P, et al. Cytogenetic study of malignant triton tumor: a case report. Cancer Genet Cytogenet 2003;144:100–5. [DOI] [PubMed] [Google Scholar]
- 10.Gil Z , Fliss DM, Voskoboimik N, et al. Two novel translocations, t(2;4)(q35;q31) and t(X;12)(q22;q24), as the only karyotypic abnormalities in a malignant peripheral nerve sheath tumor of the skull base. Cancer Genet Cytogenet 2003;145:139–43. [DOI] [PubMed] [Google Scholar]
- 11.Mawrin C , Kirches E, Boltze C, et al. Immunohistochemical and molecular analysis of p53, RB, and PTEN in malignant peripheral nerve sheath tumors. Virchows Arch 2002;440:610–15. [DOI] [PubMed] [Google Scholar]
- 12.Scheithauer BW, Woodruff JM, Erlandson RA. Primary malignant tumors of peripheral nerve. In: Tumors of the peripheral nervous system. Atlas of tumor pathology, 3rd series, fascicle 24. Washington, DC: Armed Forces Institute of Pathology, 1999:303–72.
- 13.Mitelman F, ed. ISCN: an international system for human cytogenetic nomenclature. Basel: Karger, 1995.
- 14.Safar A , Nelson M, Neff JR, et al. Recurrent anomalies of 6q25 in chondromyxoid fibroma. Hum Pathol 2000;31:306–11. [DOI] [PubMed] [Google Scholar]
- 15.Safar A , Wickert R, Nelson M, et al. Characterization of a variant SYT/SSX1 synovial sarcoma fusion transcript. Mol Diagn Pathol 1998;7:283–7. [DOI] [PubMed] [Google Scholar]
- 16.Bernards A . Neurofibromatosis type 1 and Ras-mediated signaling: filling in the GAPs. Biochim Biophys Acta 1995;1242:43–59. [DOI] [PubMed] [Google Scholar]
- 17.Shen MH, Harper PS, Upadhyaya M. Molecular genetics of neurofibromatosis type 1 (NF1). J Med Genet 1996;33:2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doorn P , Molenar W, Buter J, et al. Malignant peripheral nerve sheath tumors in patients with and without neurofibromatosis. Eur J Surg Oncol 1995;21:78–82. [DOI] [PubMed] [Google Scholar]
- 19.Bridge JA, Sandberg AA. Cytogenetic and molecular genetic techniques as adjunctive approaches in the diagnosis of bone and soft tissue tumors. Skeletal Radiol 2000;29:249–58. [DOI] [PubMed] [Google Scholar]
- 20.Jeon I-S, Davis JN, Braun BS, et al. A variant Ewing’s sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene 1995;10:1229–34. [PubMed] [Google Scholar]
- 21.Nicolaides NC, Papadopoulos N, Liu B, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 1994;371:75–80. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton SR, Liu B, Parsons RE, et al. The molecular basis of Turcot’s syndrome. N Engl J Med 1995;332:839–47. [DOI] [PubMed] [Google Scholar]
- 23.LeBeau MM, Westbrook CA, Diaz MO, et al. Evidence for two distinct c-src loci on human chromosomes 1 and 20. Nature 1984;312:70–1. [DOI] [PubMed] [Google Scholar]
- 24.Irby RB, Mao W, Coppola D, et al. Activating SRC mutation in a subset of advanced human colon cancers. Nat Genet 1999;21:187–90. [DOI] [PubMed] [Google Scholar]
- 25.Rouleau GA, Merel P, Lutchman M, et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature 1993;363:515–21. [DOI] [PubMed] [Google Scholar]
- 26.Rey JA, Bello MJ, Kusak ME, et al. Involvement of 22q12 in a neurofibrosarcoma in neurofibromatosis type I. Cancer Genet Cytogenet 1993;66:28–32. [DOI] [PubMed] [Google Scholar]
- 27.Perry A , Kunz SN, Ruller CE, et al. Differential NF1, p16, and EGFR patterns by interphase cytogenetics (FISH) in malignant peripheral nerve sheath tumor (MPNST) and morphologically similar spindle cell neoplasms. J Neuropathol Exp Neurol 2002;61:702–9. [DOI] [PubMed] [Google Scholar]
- 28.Kourea HP, Orlow I, Scheithauer BW, et al. Deletion of the INK4a gene occurs in malignant peripheral nerve sheath tumors but not in neurofibromas. Am J Pathol 1999;155:1855–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen GP, Stemmer-Rachmaninov AO, Ino Y, et al. Malignant transformation of neurofibromas in neurofibromatosis 1 is associated with CDKN2A/p16 inactivation. Am J Pathol 1999;155:1879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leguis E , Marchuk DA, Collins FS, et al. Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumour suppressor gene hypothesis. Nat Genet 1993;3:122–6. [DOI] [PubMed] [Google Scholar]
- 31.Menon AG, Anderson KM, Riccardi VM, et al. Chromosome 17p deletions and p53 gene mutations associated with the formation of malignant neurofibrosarcomas in von Recklinghausen neurofibromatosis. Proc Natl Acad Sci U S A 1990;87:5435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Legius E , Dierrick H, Wu R, et al. TP53 mutations are frequent in malignant NF1 tumors. Genes Chromosomes Cancer 1994;10:250–5. [DOI] [PubMed] [Google Scholar]
- 33.Lothe RA, Slettan A, Saeter G, et al. Alterations at chromosome 17 loci in peripheral nerve sheath tumors. J Neuropathol Exp Neurol 1995;54:65–73. [DOI] [PubMed] [Google Scholar]
- 34.Weeber EJ, Sweatt JD. Molecular neurobiology of human cognition. Neuron 2002;33:845–8. [DOI] [PubMed] [Google Scholar]
- 35.Saito T , Seki N, Matsuda Y, et al. Identification of the human ERK gene as a putative receptor tyrosine kinase and its chromosomal localization to 1p36. 1: a comparative mapping of human, mouse, and rat chromosomes, Genomics 1995;26:382–4. [DOI] [PubMed] [Google Scholar]
- 36.O’Sullivan MJ, Kyriakos M, Zhu X, et al. Malignant peripheral nerve sheath tumors with t(X;18). A pathologic and molecular genetic study. Mod Pathol 2000;13:1253–63. [DOI] [PubMed] [Google Scholar]