Abstract
Aims: To analyse the expression of proteins involved in DNA double strand break detection and repair in the luminal and myoepithelial compartments of benign breast lesions and malignant breast tumours with myoepithelial differentiation.
Methods: Expression of the ataxia telangiectasia (ATM) and p53 proteins was immunohistochemically evaluated in 18 benign and malignant myoepithelial tumours of the breast. Fifteen benign breast lesions with prominent myoepithelial compartment were also evaluated for these proteins, in addition to those in the MRE11–Rad50–NBS1 (MRN) complex, and the expression profiles were compared with those seen in eight independent non-cancer (normal breast) samples and in the surrounding normal tissues of the benign and malignant tumours examined.
Results: ATM expression was higher in the myoepithelial compartment of three of 15 benign breast lesions and lower in the luminal compartment of eight of these lesions compared with that found in the corresponding normal tissue compartments. Malignant myoepithelial tumours overexpressed ATM in one of 18 cases. p53 was consistently negative in benign lesions and was overexpressed in eight of 18 malignant tumours. In benign breast lesions, expression of the MRN complex was significantly more reduced in myoepithelial cells (up to 73%) than in luminal cells (up to 40%) (p = 0.0005).
Conclusions: Malignant myoepithelial tumours rarely overexpress ATM but are frequently positive for p53. In benign breast lesions, expression of the MRN complex was more frequently reduced in the myoepithelial than in the luminal epithelial compartment, suggesting different DNA repair capabilities in these two cell types.
Keywords: breast, myoepithelial cells, DNA repair, immunohistochemistry
The normal breast is composed of two distinct epithelial cell types, the inner or luminal secretory epithelial cells, which are surrounded by an incomplete outer layer of contractile myoepithelial cells.1–3 The identification of myoepithelial cells is important in routine histopathological diagnosis to distinguish preinvasive from invasive malignant breast lesions.2 Owing to their dual (epithelial and smooth muscle) properties, myoepithelial cells can be identified in tissue sections through a combination of immunohistochemical markers related either to the basal epithelial phenotype (expression of high molecular weight cytokeratins (CKs), namely CK5/6 and CK14) or smooth muscle contractile apparatus (α smooth muscle actin (αSMA), calponin, caldesmone, or smooth muscle myosin heavy chain).2–4 Recently, other markers that are relatively specific for myoepithelial cells but not related to these properties have been described, including p63, maspin, and neuropilin 1.2,3,5,6
“Myoepithelial cells appear to transform infrequently and tumours with myoepithelial differentiation are rare in clinical practice”
Despite the fact that breast ducts and lobules are composed of an approximately equal proportion of luminal and myoepithelial cells, myoepithelial cells appear to transform infrequently and tumours with myoepithelial differentiation are rare in clinical practice.7 Previous work on myoepithelial carcinomas has revealed few genetic alterations in these tumours compared with invasive ductal carcinomas of no special type (mean numbers: 2.1 and 8.6, respectively),8,9 suggesting that the different frequency and biology of these tumours may be reflected at the molecular genetic level.
Our previous work investigating the expression of proteins involved in the detection and repair of DNA damage, including ataxia telangiectasia mutated (ATM), p53, and the MRE11–Rad50–NBS1 (MRN) complex, have found differential expression of these proteins in normal myoepithelial and luminal breast epithelial cells.10,11 In brief, p53 immunoreactivity was negative in both cell types, whereas the other proteins showed nuclear staining of moderate to normal intensity in at least 75% of the luminal breast epithelial cells. The expression profiles in the myoepithelial compartment showed a greater variability, with low expression of the ATM protein and moderate expression of the MRN complex in > 75% of cells. Interestingly, Clarke et al reported upregulation of the ATM protein in the myoepithelial compartment of sclerosing adenosis, and suggested that the transcription of the ATM gene was upregulated in response to molecular changes associated with cell proliferation in this cell type.12
To characterise further the expression of proteins involved in DNA double break strand repair in proliferating breast cells, we investigated the differential expression of ATM, p53, and the MRN complex in both the luminal and myoepithelial compartments of 15 benign breast lesions. In addition, we assessed the expression of ATM and p53 in malignant myoepithelial tumours of the breast to determine whether their expression profile resembled that seen in malignant luminal epithelial cells or that reported for proliferating myoepithelial cells
MATERIALS AND METHODS
Patient samples
Normal breast samples from reduction mammoplasties (n = 8), sclerosing adenosis (n = 8), and breast papillomas (n = 7) were retrieved from the pathology files of the Royal Marsden Hospital (London, UK) or the Centre Léon Bérard (Lyon, France). Myoepithelial tumours—three adenomyoepitheliomas, 10 myoepithelial carcinomas, and five metaplastic breast carcinomas—were retrieved from the consultation files of one of the authors (SRL). The diagnostic criteria for myoepithelial tumours are those used in previous publications by the authors.8,13–15
Immunohistochemical analysis
Immunohistochemistry was carried out using antibodies validated in previously described protocols.10,11 Briefly, slides were dewaxed and endogenous peroxidase activity was inactivated by incubation for 30 minutes in 0.3% H2O2/methanol. For all the proteins studied, except p53, sections were treated with antigen unmasking solution (Vector Laboratories, Peterborough, UK) for 3 × 5 minutes in a microwave oven, according to the supplier’s specifications. After blocking the non-specific sites with phosphate buffered saline containing 0.1% bovine serum albumin for 45 minutes, the primary antibodies were incubated overnight at 4°C: ATML2p for ATM immunostaining (1/500 dilution; epitope amino acid 2581–2599; generously provided by Dr Y Shiloh, Tel Aviv University, Ramat Aviv, Israel) and CM1 for p53 detection (1/500 dilution; wild-type and mutant forms of human p53 protein; Tebu International, Le Perray en Yvelines, France). The expression of the Rad50 protein was analysed using the Rad50–13B3 antibody (1/100 dilution epitope amino acid 1–425; GeneTex, San Antonio, USA) and the NBS1 (Nijmegen breakage syndrome) and MRE11 (meiotic recombination 11 homologue) proteins were detected using polyclonal antibodies generously provided by Dr J Petrini (Memorial Sloan Kettering Cancer Center, New York, USA) diluted at 1/30 000 and 1/50 000, respectively. The antimouse (for ATM and Rad50 staining) or the antirabbit (for p53, NBS1, and MRE11 staining) biotinylated secondary antibody was added to the sections for 45 minutes and the peroxidase reaction was revealed using a diaminobenzidine substrate (Vector Laboratories), following streptavidin signal amplification (Vectastain ABC kit; Vector Laboratories). Anti-p63 (clone 4A4; 1/200 dilution; Santa Cruz Biotechnology, Santa Crux, California, USA), anti-CK5/6 (clone D5/16B4; 1/400 dilution; Chemicon, Hampshire, UK), anti-αSMA (clone 1A4; 1/50 dilution; Dako, Glostrup, Denmark) were used for the identification of myoepithelial cells. The expression of the α oestrogen receptor (clone 1D5; 1/40 dilution; Dako) was also assessed in the myoepithelial tumours. Positive and negative controls were included in each slide run and gave appropriate results. The cellular localisation, the staining intensity, and the percentage of tumour cells with positive nuclear staining were assessed by four pathologists. In each series of samples, normal tissues adjacent to the lesions were analysed for comparison.
Immunohistochemical scoring
For the ATM and MRN complex, expression was evaluated using a previously described scoring system.11 Briefly, nuclear staining intensity was graded as null (0), low (1), moderate (3), and high (5) compared with the immunoreactivity seen in the adjacent normal luminal epithelial compartment for the same antibody. The percentage of cells with positive nuclear staining was categorised as follows: 1, < 25%; 2, 25–50%; 3, 50–75%; and 4, > 75%, and the resulting total scores (from 0 to a maximum of 20) allowed the classification of the expression of each protein as either negative (score, 0), low (score, 1–5), intermediate (score, 6–12), or the same as that seen in the corresponding normal luminal epithelial compartment (score, > 12). Relative protein expression in the myoepithelial compartments was compared with that seen in the luminal compartments. In all benign lesions, both the luminal epithelial and the myoepithelial compartments were assessed individually. p53 expression was considered positive when more than 10% of the tumour cells showed nuclear p53 staining.
RESULTS
In normal breast tissues ATM, MRE11, Rad50, and NBS1 were highly expressed in the nuclear compartment of luminal epithelial cells (fig 1). 10,11 In myoepithelial cells there was moderate to high nuclear staining for MRE11, Rad50, and NBS1, whereas only low expression of ATM was seen, as reported previously.10,11 No cytoplasmic staining was seen in the control samples. p53 immunoreactivity was negative in both the luminal and myoepithelial cells in all the control samples.
Figure 1.
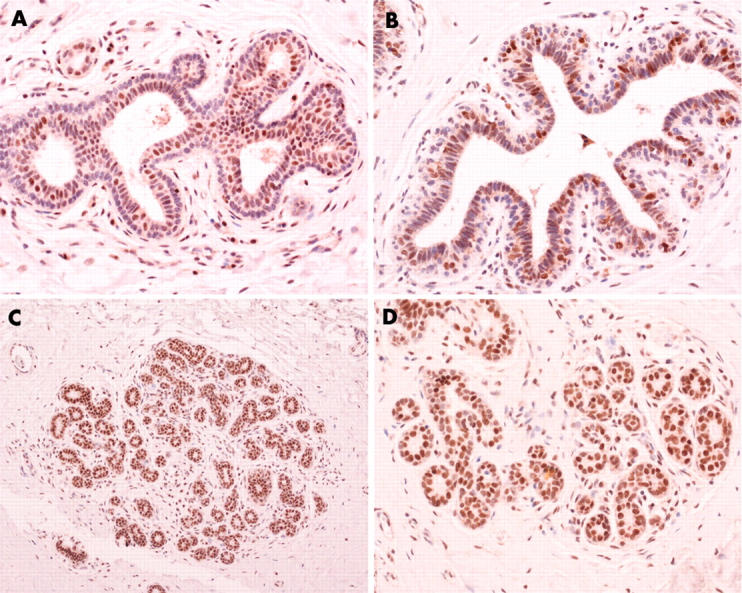
Expression of DNA double strand break repair proteins in normal breast tissue: (A) ATM, (B) NBS1, (C) MRE11, (D) Rad50. (Original magnification: A, B, D, ×200; C, ×100.)
Increased expression of ATM was seen in the p63, αSMA, and CK5/6 positive myoepithelial cells of three of the seven papillomas (fig 2) but in none of the cases of sclerosing adenosis (fig 3). In the luminal cells, ATM expression appears to be reduced (score, < 5) in four of the eight sclerosing adenosis and four of the seven papillomas analysed. p53 overexpression was not seen in these benign lesions of the breast. Reduced expression of NBS1, MRE11, and Rad50 was observed in the myoepithelial compartment of 11, nine, and five of the 15 benign breast lesions, respectively. These proteins were reduced in the luminal compartment of six, three, and two of these same lesions, respectively (table 1). When taken together, a significant difference between the expression of the proteins in the MRN complex in the luminal and myoepithelial compartments was seen in these benign lesions (p = 0.0005; Fisher’s exact test).
Figure 2.
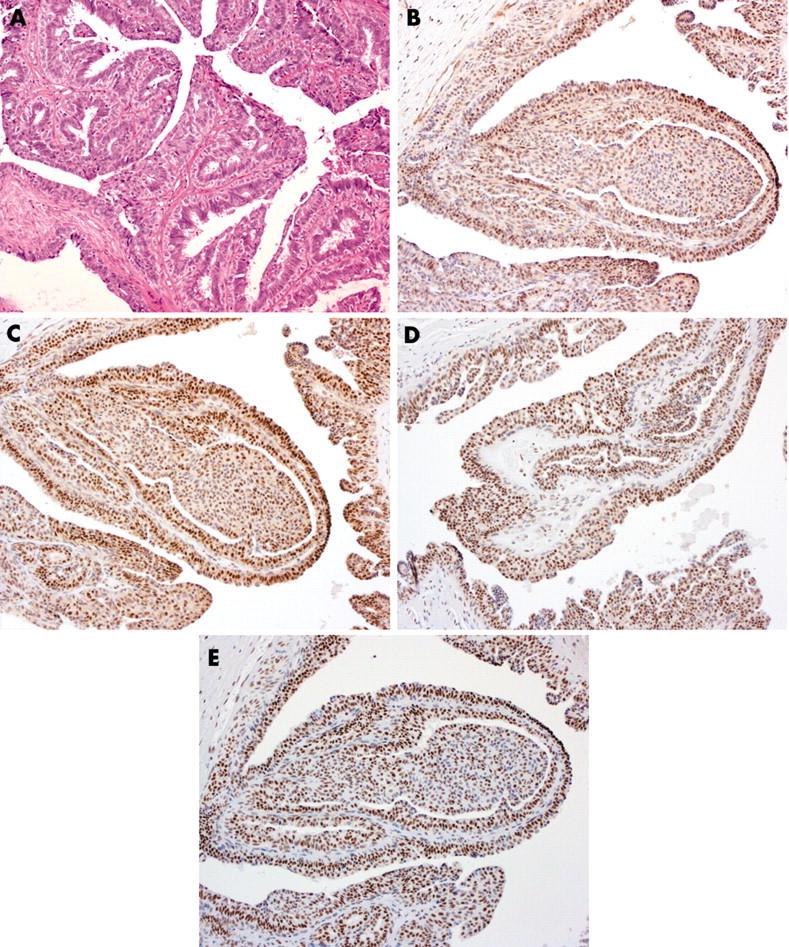
Expression of DNA double strand break repair proteins in a benign papilloma: (A) haematoxylin and eosin, (B) ATM, (C) NBS1, (D) MRE11, (E) Rad50 (original magnification, ×200).
Figure 3.
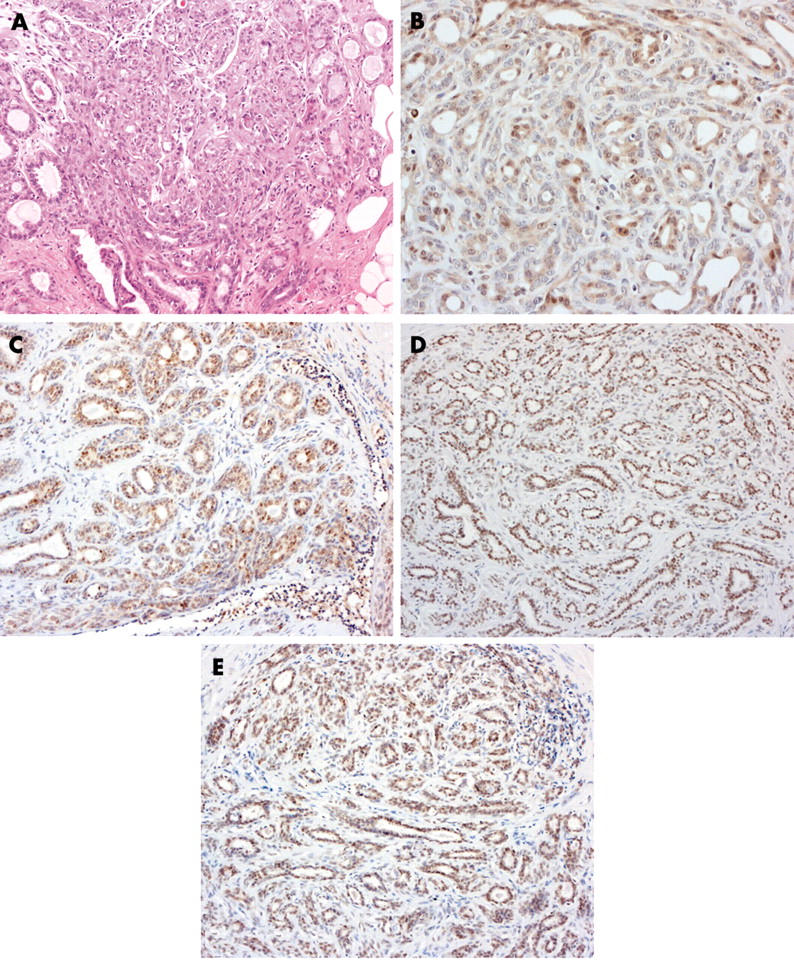
Expression of DNA double strand break repair proteins in a case of sclerosing adenosis: (A) haematoxylin and eosin, (B) ATM, (C) NBS1, (D) MRE11, (E) Rad50. (Original magnification: C, D, E, ×100; A, B, ×200.)
Table 1.
Expression of ATM, p53, and NBS1–MRE11-Rad50 complex proteins in benign breast lesions
| Case | Diagnosis | ATM | p53 | NBS1 score | MRE11 score | Rad50 score | ||||
| Myo | Lum | Myo | Lum | Myo | Lum | Myo | Lum | |||
| 1 | Papilloma | 1 | 1 | Negative | 1 | 1 | 20 | 20 | 20 | 20 |
| 2 | Papilloma | 1 | 3 | Negative | 6 | 12 | 15 | 12 | 9 | 12 |
| 3 | Papilloma | 1 | 2 | Negative | 1 | 3 | 3 | 12 | 3 | 9 |
| 4 | Papilloma | 2 | 4 | Negative | 3 | 20 | 1 | 20 | 1 | 12 |
| 5 | Papilloma | 9 | 9 | Negative | 3 | 12 | 3 | 12 | 9 | 12 |
| 6 | Papilloma | 9 | 12 | Negative | 15 | 20 | 15 | 20 | 15 | 20 |
| 7 | Papilloma | 12 | 12 | Negative | 12 | 12 | 3 | 1 | 9 | 12 |
| 8 | Sclerosing adenosis | 0 | 3 | Negative | 1 | 1 | 1 | 1 | 1 | 4 |
| 9 | Sclerosing adenosis | 1 | 1 | Negative | 2 | 3 | 2 | 3 | 2 | 3 |
| 10 | Sclerosing adenosis | 1 | 3 | Negative | 1 | 12 | 6 | 12 | 6 | 12 |
| 11 | Sclerosing adenosis | 1 | 3 | Negative | 1 | 3 | 3 | 12 | 6 | 20 |
| 12 | Sclerosing adenosis | 3 | 9 | Negative | 12 | 20 | 20 | 20 | 20 | 20 |
| 13 | Sclerosing adenosis | 3 | 20 | Negative | 3 | 12 | 6 | 20 | 6 | 12 |
| 14 | Sclerosing adenosis | 3 | 9 | Negative | 0 | 4 | 3 | 12 | 6 | 12 |
| 15 | Sclerosing adenosis | 3 | 20 | Negative | 3 | 6 | 3 | 20 | 3 | 20 |
Myo, myoepithelial cells; Lum, luminal epithelial cells.
Owing to limited amounts of material, only p63 (n = 16), αSMA (n = 13), CK5/6 (n = 12), p53 (n = 18), and ATM (n = 18) were studied in breast tumours with myoepithelial differentiation. p63 was positive in two of two adenomyoepitheliomas, nine of 10 myoepithelial carcinomas, and four of four metaplastic breast carcinomas. αSMA was positive in one of one adenomyoepithelioma, four of six myoepithelial carcinomas, and two of five metaplastic breast carcinomas. CK5/6 was positive in one of one adenomyoepithelioma, eight of eight myoepithelial carcinomas, and four of four metaplastic breast carcinomas. Only one of one adenomyoepithelioma and one of six malignant myoepitheliomas were positive for oestrogen receptor. Low expression of ATM was seen in 17 of the 18 neoplasms, whereas p53 was positive in eight (fig 4; table 2).
Figure 4.
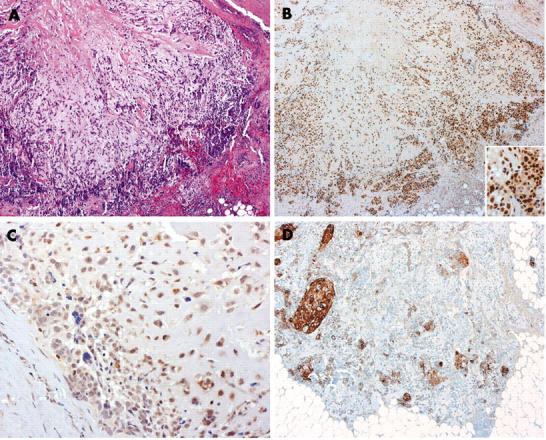
Matrix producing breast carcinoma with focal squamous differentiation. (A) haematoxylin and eosin, (B) p53, (C) ATM, (D) cytokeratin 5/6. (Original magnification: A, B, ×4; D, ×100; inset of B, C, ×200.)
Table 2.
Expression of ATM, p53, and MRN complex proteins in myoepithelial tumours
| Case | Diagnosis | ATM score | p53 | p63 | CK5/6 | ER | αSMA |
| 1 | Adenomyoepithelioma | 4 | Negative | + | + | NA | NA |
| 2 | Adenomyoepithelioma | 0 | Negative | NA | NA | NA | NA |
| 3 | Adenomyoepithelioma | 0 | NA | ++ | NA | ++ | ++ |
| 4 | Malignant myoepithelioma | 3 | Positive | + | + | – | + |
| 5 | Malignant myoepithelioma | 3 | Negative | ++ | ++ | NA | NA |
| 6 | Malignant myoepithelioma | 2 | Positive | ++ | ++ | – | ++ |
| 7 | Malignant myoepithelioma | 2 | Positive | ++ | ++ | – | ++ |
| 8 | Malignant myoepithelioma | 0 | Positive | ++ | ++ | – | + |
| 9 | Malignant myoepithelioma | 0 | Positive | – | NA | NA | NA |
| 10 | Malignant myoepithelioma | 0 | Negative | ++ | ++ | – | – |
| 11 | Malignant myoepithelioma | 0 | Negative | ++ | ++ | NA | NA |
| 12 | Malignant myoepithelioma | 0 | Negative | ++ | NA | NA | NA |
| 13 | Malignant myoepithelioma | 0 | NA | ++ | ++ | ++ | – |
| 14 | Matrix producing breast carcinoma | 3 | Positive | + | + | – | + |
| 15 | Sarcomatoid breast carcinoma | 0 | Positive | + | + | – | ++ |
| 16 | Sarcomatoid breast carcinoma | 0 | Negative | NA | NA | – | – |
| 17 | Sarcomatoid breast carcinoma | 12 | Positive | ++ | + | – | – |
| 18 | Sarcomatoid breast carcinoma | 0 | Negative | ++ | ++ | – | – |
+, focally positive; ++, diffusely positive; αSMA, α smooth muscle actin; ER, oestrogen receptor; NA, not available.
DISCUSSION
The ATM protein and the MRN complex belong to the BASC (BRCA1 associated genome surveillance complex) complex, which includes other proteins such as BRCA1, BLM, hMSH2, hMSH6, and hMLH1.16 This complex is reported to play a role in the recognition of DNA damage and the activation of signalling pathways leading to cell cycle checkpoint activation, DNA repair, or apoptosis. In response to DNA damage, the protein kinase ATM is activated by autophosphorylation and phosphorylates several proteins such as p53, Mdm2, Chk2, BRCA1, NBS1, c-abl, SMC1, BLM, and FancD2, which are involved in genome integrity maintenance (reviewed in Shiloh17). The abnormal expression of such proteins in breast tissue could result in increased genomic instability and influence mammary tumorigenesis (reviewed in Angèle and Hall18).
ATM, MRE11, Rad50, and NBS1 show differential expression in normal breast tissue with respect to cell type.10,11 All four proteins are moderately to strongly expressed in the inner luminal cells. In the outer myoepithelial cells, MRE11, Rad50, and NBS1 are also highly expressed, whereas the expression of ATM is weak.
Our investigation of benign breast lesions showing a distinct myoepithelial compartment revealed low ATM expression in the myoepithelial cells in four of seven papillomas and in all eight cases of sclerosing adenosis. ATM expression was reduced in luminal cells in approximately 50% of these benign lesions. This low ATM expression seen in all the sclerosing adenosis cases studied here is in contrast to the results of Clarke et al,12 who reported strong nuclear and cytoplasmic ATM immunoreactivity in myoepithelial cells of six of seven and four of seven cases of sclerosing adenosis, respectively. The reason for this difference in ATM expression is unclear. However, it should be noted that Clarke and colleagues12 used a different anti-ATM antibody that revealed a high degree of cytoplasmic ATM expression, which we did not see in normal tissues, and was positive in only one of 99 ductal carcinomas of the breast examined.11
In our present study, we have analysed 18 breast tumours with a myoepithelial phenotype. ATM does not appear to play a major role in the carcinogenesis of these lesions because only one of 18 of these tumours showed ATM expression and expression was very low in normal myoepithelial cells. This is in contrast to ductal breast carcinomas, in which reduced ATM expression, at both the mRNA and protein level, has been reported in up to 75% of tumours compared with normal luminal epithelial cells.10,11,19,20 Interestingly, immunohistochemical expression of p53 was detected in 44% of malignant myoepithelial tumours. Unfortunately, owing to restraints in tissue availability it was not possible to determine the underlying alterations in the TP53 gene associated with this p53 protein overexpression.
“ATM does not appear to play a major role in the carcinogenesis of breast tumours with a myoepithelial phenotype”
Expression of the MRN complex in breast tumours has been investigated in only a few studies. We found reduced expression of the protein complex in 28–46 of 99 cases of invasive breast ductal carcinoma, frequently in association with an alteration of ATM or p53 expression,11 whereas Sakata and colleagues21 reported normal expression of NBS1 in 10 breast adenocarcinomas. Unfortunately, because of limited amounts of material, we could not investigate the expression of the MRN complex in the rare malignant myoepithelial lesions. However, we did investigate these proteins in the benign papillomas and sclerosing adenosis. We found that a reduction in MRN complex proteins was significantly more prevalent in the myoepithelial compartment of these lesions, compared with the luminal compartment, with a good correlation in the degree of expression of the three proteins, as reported previously.11
Taken together, these results suggest that the two epithelial cell types of the breast show a different profile of DNA repair protein expression. This may reflect different mechanisms of tumorigenesis and/or different degrees of genomic instability.8,9 Detailed functional studies will be required to investigate this issue further.
Take home messages.
Malignant myoepithelial tumours rarely overexpress ATM (ataxia telangiectasia mutated), a protein involved in DNA double strand break detection and repair, but are frequently positive for p53
In benign breast lesions, expression of the MRE11–Rad50–NBS1 complex (also involved in DNA double strand break detection and repair) was more frequently reduced in the myoepithelial than in the luminal epithelial compartment, suggesting different DNA repair capabilities in these two cell types
This may reflect different mechanisms of tumorigenesis and/or different degrees of genomic instability in these two cell types
Acknowledgments
We are grateful to Dr Y Shiloh, Dr J Petrini, and the GeneTex Company for their generous gifts of antibodies. The technical assistance of C Carreira, S Roche, and N Lyandrat from the common service of immunohistochemistry is also gratefully acknowledged. J S Reis-Filho is the recipient of the Gordon Signy International Fellowship Award of the World Association of Societies of Pathology and Laboratory Medicine (WASPALM) and is partially supported by a PhD grant (reference: SFRH/BD/5386/2001) from the Fundação para a Ciência e a Tecnologia (FCT), Portugal. Financial support through the Emergence Programme (2002) of the Rhône-Alpes Region of France is gratefully acknowledged.
Abbreviations
αSMA, α smooth muscle actin
ATM, ataxia telangiectasia mutated
CK, cytokeratin
ER, oestrogen receptor
MRN, complex of MRE11, Rad50 and NBS1 proteins
MRE11, meiotic recombination 11 homologue
NBS, Nijmegen breakage syndrome
REFERENCES
- 1.Moritani S , Kushima R, Sugihara H, et al. Availability of CD10 immunohistochemistry as a marker of breast myoepithelial cells on paraffin sections. Mod Pathol 2002;15:397–405. [DOI] [PubMed] [Google Scholar]
- 2.Yaziji H , Gown AM, Sneige N. Detection of stromal invasion in breast cancer: the myoepithelial markers. Adv Anat Pathol 2000;7:100–9. [DOI] [PubMed] [Google Scholar]
- 3.Reis-Filho JS, Schmitt FC. Taking advantage of basic research: p63 is a reliable myoepithelial and stem cell marker. Adv Anat Pathol 2002;9:280–9. [DOI] [PubMed] [Google Scholar]
- 4.Foschini MP, Scarpellini F, Gown AM, et al. Differential expression of myoepithelial markers in salivary, sweat and mammary glands. Int J Surg Pathol 2000;8:29–37. [DOI] [PubMed] [Google Scholar]
- 5.Barbareschi M , Pecciarini L, Cangi MG, et al. p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am J Surg Pathol 2001;25:1054–60. [DOI] [PubMed] [Google Scholar]
- 6.Stephenson JM, Banerjee S, Saxena NK, et al. Neuropilin-1 is differentially expressed in myoepithelial cells and vascular smooth muscle cells in preneoplastic and neoplastic human breast: a possible marker for the progression of breast cancer. Int J Cancer 2002;101:409–14. [DOI] [PubMed] [Google Scholar]
- 7.Lakhani SR, O’Hare MJ. The mammary myoepithelial cell—Cinderella or ugly sister? Breast Cancer Res 2001;3:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones C , Foschini MP, Chaggar R, et al. Comparative genomic hybridization analysis of myoepithelial carcinoma of the breast. Lab Invest 2000;80:831–6. [DOI] [PubMed] [Google Scholar]
- 9.Jones C , Nonni AV, Fulford L, et al. CGH analysis of ductal carcinoma of the breast with basaloid/myoepithelial cell differentiation. Br J Cancer 2001;85:422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angèle S , Treilleux I, Taniere P, et al. Abnormal expression of the ATM and TP53 genes in sporadic breast carcinomas. Clin Cancer Res 2000;6:3536–44. [PubMed] [Google Scholar]
- 11.Angèle S , Treilleux I, Brémond A, et al. Altered expression of DNA double-strand break detection and repair proteins in breast carcinomas. Histopathology 2003;43:347–53. [DOI] [PubMed] [Google Scholar]
- 12.Clarke RA, Kairouz R, Watters D, et al. Upregulation of ATM in sclerosing adenosis of the breast. Mol Pathol 1998;51:224–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foschini MP, Eusebi V. Carcinomas of the breast showing myoepithelial cell differentiation. A review of the literature. Virchows Arch 1998;432:303–10. [DOI] [PubMed] [Google Scholar]
- 14.Foschini MP, Reis-Filho JS, Eusebi V, et al. Salivary gland-like tumours of the breast: surgical and molecular pathology. J Clin Pathol 2003;56:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reis-Filho JS, Milanezi F, Paredes J, et al. Novel and classic myoepithelial/stem cell markers in metaplastic carcinomas of the breast. Appl Immunohistoche Mol Morphol 2003;11:1–8. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y , Cortez D, Yazdi P, et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 2000;14:927–39. [PMC free article] [PubMed] [Google Scholar]
- 17.Shiloh Y . ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 2003;3:155–68. [DOI] [PubMed] [Google Scholar]
- 18.Angèle S , Hall J. The ATM gene and breast cancer: is it really a risk factor? Mutat Res 2000;462:167–78. [DOI] [PubMed] [Google Scholar]
- 19.Waha A , Sturne C, Kessler A, et al. Expression of the ATM gene is significantly reduced in sporadic breast carcinomas. Int J Cancer 1998;78:306–9. [DOI] [PubMed] [Google Scholar]
- 20.Kairouz R , Clarke RA, Marr PJ, et al. ATM protein synthesis patterns in sporadic breast cancer. Mol Pathol 1999;52:252–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakata K , Matsumoto Y, Tauchi H, et al. Expression of genes involved in repair of DNA double-strand breaks in normal and tumor tissues. Int J Radiat Oncol Biol Phys 2001;49:161–7. [DOI] [PubMed] [Google Scholar]


