Abstract
Background: The discovery that genetic alterations in oncogenes and tumour suppressor genes accompany tumour formation in many human tumours has encouraged the search for genes that promote or suppress tumour spread and metastasis; nm23 is a promising candidate for a metastasis suppressing gene.
Aims: To evaluate whether expression of nm23-H1 protein or loss of heterozygosity (LOH) of the nm23-H1 gene is associated with colon cancer progression.
Materials/Methods: Paraffin wax embedded tissue sections were analysed immunohistochemically. DNA isolated from normal and tumour tissue was used for LOH analysis using a variable nucleotide tandem repeat (VNTR) marker located in the untranslated 5′ region of the nm23-H1 gene. RNA isolated from tumour and normal tissue was used for “real time” RT-PCR.
Results: Of 102 adenocarcinomas examined, 58.8% stained weakly for nm23-H1 protein. There was a negative correlation between nm23-H1 positivity and tumour histological grade. In VNTR analysis, 70.2% of patients were informative and 27.4% of tumours had nm23-H1 LOH. There was a positive correlation between nm23-H1 LOH and both tumour histological grade and Dukes’s stage. Expression of nm23-H1 mRNA was increased in 22 of 30 colon tumours compared with normal tissue. No significant correlation was found between nm23-H1 mRNA expression and histological grade or Dukes’s stage of tumours.
Conclusions: These findings suggest that nm23-H1 protein expression in early stages may have a role in suppressing metastasis in sporadic colon cancer, whereas at a later stage both reduced nm23-H1 protein expression and LOH of the nm23-H1 gene may play role in colon cancer progression and metastasis.
Keywords: loss of heterozygosity, colon carcinoma, nm23-H1, protein expression, real time reverse transcription polymerase chain reaction
A cascade of cellular, biochemical, and genetic events is known to occur in the development and progression of tumours, leading to malignancy and ultimately to metastasis. Tumour metastasis, the process by which tumour cells leave the primary tumour to colonise other sites of the body, is a major cause of death for patients with cancer. The complexity of metastatic dissemination of tumour cells can be realised by considering the steps that they must perform before successfully colonising a distant site. Metastasising cells must first disseminate from the primary tumour, invade the surrounding tissue, intravasate and extravasate the circulatory system, arrest, initiate angiogenesis, and colonise distant sites, at the same time evading the immune system.1,2
“nm23 seemed to be the most promising candidate for a gene with metastasis suppressing function”
The discovery of genetic alterations in oncogenes and tumour suppressor genes, which accompany tumour formation in a wide variety of human tumour types, has encouraged the search for genes that may promote or suppress tumour spread and metastasis. Among these, nm23 seemed to be the most promising candidate for a gene with metastasis suppressing function.3 The nm23 gene was originally identified by differential hybridisation of K-1735 melanoma cell line clones of varying metastatic potential. A tumour metastasis suppressor function was implicated by the reduced expression of nm23 in highly metastatic sublines compared with non-metastatic sublines derived from the same K-1735 clone.4 Two human nm23 genes, nm23-H1 and nm23-H2, have been cloned.5 They are 88% homologous to each other and encode two polypeptide subunits of a nucleoside diphosphate (NDP) kinase. NDP kinase transfers the γ phosphate of nucleoside triphosphate to NDP via a high energy phosphohistidine intermediate. It has been shown, however, that the biological function of nm23 is not related to its NDP kinase activity.6 Rather, the motility suppressive function of nm23-H1 is probably associated with histidine dependent phosphotransferase activity of the molecule.7
The expression of nm23 has been shown to be raised in several different tumours of lower metastatic potential than in the corresponding tumours of higher metastatic potential, including breast, hepatocellular, ovarian, and gastric carcinomas, and melanoma.8 In other tumours, such as neuroblastoma, pancreatic carcinoma, and head and neck carcinomas, surprisingly, the opposite trend has been reported.9
The role of nm23-H1 in colorectal carcinoma is still controversial. At the protein level, several studies have reported an inverse association between nm23-H1 expression and tumour stage,10–12 whereas others have found no significant association.13,14 One study reported that overexpression of nm23-H1 in primary colorectal carcinoma could be linked to disease recurrence.15 At the DNA level, allelic deletion or mutation of the nm23-H1 gene appears to be associated with distant metastasis in some studies,16–18 but not in others.19,20
In our study, we analysed the nm23-H1 gene at the DNA, mRNA, and protein levels. The purpose of our study was to evaluate whether the expression of the nm23-H1 protein (as assessed by immunostaining) and loss of heterozygosity (LOH) of the nm23-H1 gene are associated with tumour stage and the grade of tumour differentiation. In addition, we also investigated the correlation between nm23-H1 expression and five year survival. We also examined the expression of nm23-H1 mRNA by “real time” reverse transcription polymerase chain reaction (RT-PCR).
MATERIAL AND METHODS
Patients and tissue specimens
Our retrospective study was carried out using specimens of normal colon tissue and benign and malignant colon lesions. All specimens were obtained from the Croatian human tumour bank.21 All specimens were obtained through routine surgery, and the diagnoses were established by standard diagnostic procedures and confirmed histopathologically. Staging was performed by Dukes’s staging.22,23 The patients were followed up according to a standardised protocol that included laboratory tests at one to two monthly intervals, chest radiography, ultrasonography, and computerised tomographic examination of the liver, in addition to endoscopy of the colon at one year intervals during the first three years postoperatively, and thereafter at six to 12 month intervals. Causes of death were ascertained from the medical records or necropsy (if performed). Patients who died within four weeks of radical surgery were excluded from the analysis. Deaths by other causes were censored observations from the time of death.
None of the patients underwent preoperative irradiation or chemotherapy. Our study comprised 62 men and 40 women, with an age range between 31 and 88 years (mean, 62.9).
Each specimen was routinely fixed in 10% formalin and immersed in melted paraffin wax. Sections (4 μm thick) were cut and mounted on to glycerine treated slides. Fresh samples of resected colon carcinoma (immediately adjacent to the segment of tissue that was fixed in formalin) and normal colon mucous adjacent (more than 5 cm from the tumour) to resected carcinoma were snap frozen in liquid nitrogen and stored in the human tumour bank at −80°C until further use. Before inclusion in our study, each specimen was verified by a histopathologist.
Immunohistochemical detection of nm23-H1 protein
After dewaxing in xylene, the slides were washed in phosphate buffered saline (PBS) (three times for three minutes). The endogenous peroxidase activity was quenched by 15 minutes of incubation in methanol with 3% hydrogen peroxide (Sigma Chemical Co, Deisenhofen, Germany). The fixed slides were then cleared with PBS. Non-specific binding was blocked by applying normal rabbit serum in a humidity chamber at a dilution of 1/10 for 30 minutes. The slides were blotted, and the primary mouse monoclonal antibody to human nm23-H1 (NM301 monoclonal antibody; Molecular Oncology Inc, Gaithersburg, Maryland, USA) at a concentration of 5 μg/ml was applied overnight at 4°C. Slides were washed three times in a series of PBS containing 3%, 2%, and 1% of normal human serum. Secondary antibody (rabbit antimouse; Dako, Glostrup, Denmark), diluted with PBS and normal human serum (40 μl rabbit antimouse antibody, 50 μl normal human serum, and 910 μl PBS), was applied for one hour at room temperature. Finally, peroxidase–antiperoxidase (Dako) conjugate diluted 1/100 in PBS was applied for one hour at room temperature. After washing with PBS, slides were kept in diaminobenzidine tetrahydrochloride (Sigma Chemical Co) for seven minutes (50 mg in 200 ml PBS with 25 μl of 30% H2O2), counterstained with haematoxylin for 30 seconds, immersed in saturated lithium carbonate for 30 seconds, and mounted in PBS/glycerol (1/1).
Evaluation of slides
For each slide, the entire tumour area was evaluated. The concentration of the antigen was assessed by estimating the relative visual intensity of chromogenic label, and the results were expressed as negative (−) or positive (+) staining. Each sample was assessed independently by two observers (SK and SS). There was a 95% initial agreement between them.
Controls
Control staining was performed by omitting the primary antibody. As a positive control in immunohistochemical studies, we used paraffin wax slides of the normal bronchial ciliated epithelium and cultured breast carcinoma cells MCF7.
Tumour and normal DNA
All specimens were obtained during routine surgery of patients with colon adenocarcinoma. Fresh samples of resected colon carcinoma and normal mucous stored in a human tumour bank before use in this study were verified by the histopathologist. All tumour specimens were examined by routine haematoxylin and eosin staining to determine the proportion of tumour cells in the sample (it had to be more than 80%). Control normal DNA was extracted from histologically normal colon mucous adjacent (more than 5 cm from the tumour) to the adenocarcinoma. Frozen tissue DNA extraction was performed using proteinase K digestion and phenol chloroform extraction.
Polymerase chain reaction
Dinucleotide (C-A)n repeat polymorphism analysis was carried out by PCR using a pair of primers (5′-TATGAGTTCAACTACGCACG-3′ and 5′-CTCGAGCACAGGAGCAGGTT-3′) flanking the repetitive untranslated 5′ region in the nm23-H1 gene. Repeat sequences (110–120 bp) were amplified from 200 ng genomic DNA in a reaction volume of 25 μl containing 10 pmol of each primer, 50 μM of each dNTP, and 1 U Taq Gold polymerase (Applied Biosystems). PCR reactions were carried out in a GeneAmp PCR System 2400 (Applied Biosystems) for 30 cycles. The samples were processed through 30 cycles consisting of 30 seconds at 95°C, 30 seconds at 55°C, and 45 seconds at 72°C, with a final extension at 72°C for 10 minutes. The amplification of the product was verified by agarose gel electrophoresis.
Variable nucleotide tandem repeat analysis
For variable nucleotide tandem repeat (VNTR) analysis, 5 μl aliquots of the PCR product were mixed with 3 μl loading buffer and loaded on to a 1 mm thick, 35 × 30 cm, 12% non-denaturing polyacrylamide gel. Electrophoresis was performed in 1× Tris/borate/EDTA buffer for 18 hours at 400 V, at room temperature. The gels were silver stained.
LOH analysis
LOH was defined by a visible change in the allele to allele ratio in tumours compared with matched normal tissue.
RNA extraction and reverse transcription
Total RNA was extracted from fresh samples of resected colon carcinoma and corresponding normal tissue stored in a human tumour bank using Trizol reagent (Invitrogen, Life Technologies, Carlsbad, California, USA), and 1 μg of RNA from each sample was used for subsequent reverse transcription (TaqMan, Reverse Transcription Reagents; Applied Biosystems, Foster City, California, USA), according to the manufacturer’s instructions.
“Real time” RT-PCR analysis of nm23-H1 mRNA expression
Real time RT-PCR analysis for the nm23-H1 gene and the internal house keeping gene RPLP0 was performed using an ABI PRISM 7000 sequence detection system (Applied Biosystems) and predeveloped TaqMan assay reagents—Hs00264824 for the nm23-H1 gene and Hs99999902 for RPLP0. The PCR reaction was carried out according to the manufacturer’s protocol. To compensate for inter-PCR variation, normalisation of the target gene (nm23-H1) with an endogenous control (RPLPO) was performed. The normalised amounts of target gene in the tumour tissue samples were then compared with the normalised amounts in the corresponding normal tissue samples. For this analysis the comparative Ct method (ΔΔCt) was used.
Statistical analysis
Categorical data were analysed using the χ2 test and Fisher’s exact test. The probability of survival was calculated for the different subgroups by the Kaplan–Meier method. Four week mortality was excluded from the survival curves. Significant differences were evaluated by the Mantel-Cox test. Box and whisker plots were generated in the basic module of the program Statistica. The correlations were analysed with the Wilcoxon rank sum test.
RESULTS
nm23-H1 protein expression in colorectal cancer
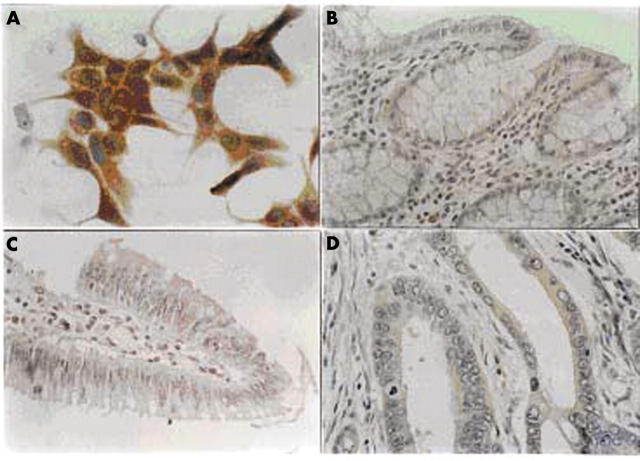
One hundred and thirty two samples (10 samples of normal colon and 122 samples of different colon neoplastic lesions) were examined for the presence of the nm23-H1 tumour suppressor protein. To verify the specificity of the monoclonal antibodies used in our experiments we tested them immunocytochemically on a human breast carcinoma cell line MCF7 (fig 1A).
Figure 1.
Immunohistochemical analysis of nm23-H1 protein expression in (A) carcinoma cell line MCF7, (B) normal colon, (C) colon adenoma, and (D) colon adenocarcinoma. The positive staining in colon adenocarcinomas was seen as weak cytoplasmic staining of tumour cells with negative surrounding tissue. Original magnification: (A, D), ×1000; (B, C), ×400).
All the histologically normal colon samples examined were negative for the nm23-H1 protein (fig 1B). All the examined hyperplastic polyps and adenomas were also negative for the nm23-H1 protein (fig 1C).
Of the 102 adenocarcinomas examined, 60 (58.8%) showed weak positive immunostaining for the nm23-H1 protein. The staining pattern was similar in all carcinomas, namely: weak cytoplasmic staining of tumour cells with negative surrounding normal tissue (fig 1D).
nm23-H1 protein expression and clinical parameters
No correlation was found between nm23-H1 protein positivity and patient age (p = 0.940; table 1). Most tumour samples from female patients were positive for the nm23-H1 protein (72.5%). There was a significant difference between the male and female groups of patients according to positivity for the nm23-H1 protein (p = 0.024; table 1). No correlation was found between nm23-H1 protein expression and the size, histological grade, or Dukes’s stage of tumours (table 1). nm23-H1 positive tumours were most frequently Dukes’s stage B (30 of 45 tested) and well differentiated (11 of 17 tested). A negative correlation was found between nm23-H1 positivity and histological grade of the tumours, although this difference was not significant (p = 0.667).
Table 1.
Clinicopathological features of 102 patients with colorectal cancer stratified by nm23-H1 status
| Characteristic | nm23-H1 staining | p Value | |
| Positive (%) (n = 60) | Negative (%) (n = 42) | ||
| Age | |||
| <70 years | 41 (60.3) | 27 (39.7) | |
| ⩾70 years | 19 (55.9) | 15 (44,1) | 0.940 |
| Sex | |||
| Male | 31 (50) | 31 (50) | |
| Female | 29 (72.5) | 11 (27.5) | 0.024 |
| Tumour size | |||
| ⩽5 cm | 37 (52.9) | 33 (47.1) | |
| >5 cm | 23 (71.9) | 9 (28.1) | 0.085 |
| Histological grade (differentiation) | |||
| Well | 11 (64.7) | 6 (35.3) | |
| Moderate | 31 (60.8) | 20 (39.2) | |
| Poor | 18 (53) | 16 (47) | 0.667 |
| Dukes’s stage | |||
| A | 15 (51.7) | 14 (48.3) | |
| B | 30 (66.7) | 15 (33.3) | |
| C | 15 (53.6) | 13 (46.4) | 0.356 |
Survival analysis
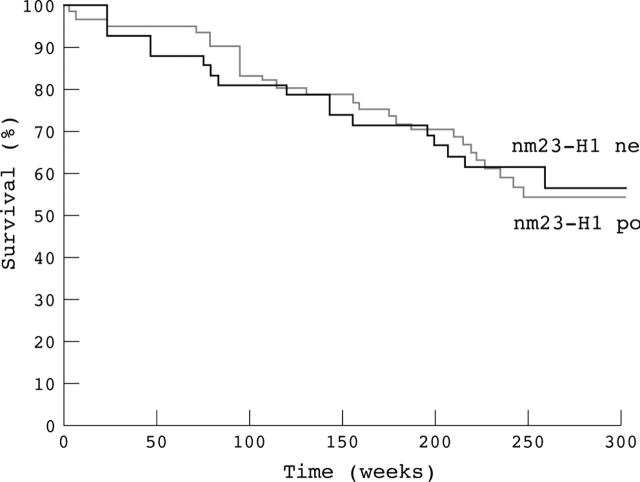
An exploratory analysis was conducted to correlate the outcome of patients monitored during the 260 week period with nm23-H1 immunoreactivity results. Survival analysis was performed on 102 patients who survived for more than four weeks after surgery. Figure 2 shows the survival curves according to nm23-H1 positivity. The median survival time of the patients without nm23-H1 protein expression was 220.3 weeks, with 56.4% of them being alive at the end of the monitoring period. The median survival time of the patients with nm23-H1 oncoprotein expression was 201.9 weeks, with 54.1% of them being alive at the end of the monitoring period. Statistical analysis showed that there was no significant difference (p = 0.712) between these two groups of patients according to nm23-H1 expression (table 2).
Figure 2.
Kaplan–Meier curves of nm23-H1 negative and nm23-H1 positive colon adenocarcinomas.
Table 2.
Survival of 102 patients with colorectal cancer stratified by clinicopathological features and nm23-H1 staining
| Characteristic | Number of cases | 5 year survival (%) | Regression analysis | |
| Relative risk of death | p Value | |||
| Age | ||||
| <70 years | 68 | 58.82 | 1 | |
| ≥70 years | 34 | 50.00 | 1.32 | 0.378 |
| Sex | ||||
| Male | 62 | 53.23 | 1 | |
| Female | 40 | 60.00 | 1.19 | 0.577 |
| Tumour size | ||||
| ⩽5 cm | 70 | 62.86 | 1 | |
| >5 cm | 32 | 40.63 | 1.74 | 0.072 |
| Histological grade (differentiation) | ||||
| Well | 17 | 82.35 | 1 | |
| Moderate | 51 | 62.75 | 2.18 | |
| Poor | 34 | 32.35 | 5.71 | 0.0055 |
| Dukes’s stage | ||||
| A | 29 | 86.21 | 1 | |
| B | 45 | 60.00 | 3.13 | |
| C | 28 | 17.86 | 10.83 | 0.00001 |
| nm23-H1 staining | ||||
| Absent | 42 | 54.76 | 1 | |
| Present | 60 | 56.67 | 1.12 | 0.712 |
LOH at the nm23-H1 gene locus
To analyse LOH at the nm23-H1 gene we used a VNTR marker located in the untranslated 5′ region of the nm23-H1 gene. The size of the PCR products was between 110 and 120 bp. Normal DNA showed one (homozygous, not informative patients) or two (heterozygous, informative patients) bands at this flanking marker. At this nm23-H1 gene locus, 73 of 104 (70.2%) patients were informative (heterozygous) and 20 of the 73 demonstrated LOH (table 3). Figure 3 shows two examples of nm23-H1 gene LOH at this locus.
Table 3.
Clinicopathological features of 73 informative patients with colorectal cancer stratified by nm23-H1 LOH status
| Characteristic | nm23-H1 LOH | ||
| Negative (%) (n = 53) | Positive (%) (n = 20) | p Value | |
| Age | |||
| <70 years | 28 (70.0) | 12 (30.0) | |
| ⩾70 years | 22 (73.3) | 8 (26.7) | 0.79 |
| Sex | |||
| Male | 20 (62.5) | 12 (37.5) | |
| Female | 33 (80.5) | 8 (19.5) | 0.11 |
| Tumour size | |||
| ⩽5 cm | 31 (73.8) | 11 (26.2) | |
| >5 cm | 11 (64.7) | 6 (35.3) | 0.53 |
| Histological grade (differentiation) | |||
| Well | 12 (80.0) | 3 (20.0) | |
| Moderate | 13 (72.2) | 5 (27.8) | |
| Poor | 7 (50.0) | 7 (50.0) | 0.24 |
| Dukes’s stage | |||
| A | 12 (80.0) | 3 (20.0) | |
| B | 12 (75.0) | 4 (25.0) | |
| C | 25 (67.6) | 12 (32.4) | 0.72 |
LOH, loss of heterozygosity.
Figure 3.
Loss of heterozygosity (LOH) at the nm23-H1 gene microsatellite locus in sporadic colon adenocarcinomas. N, normal; T, tumour; lanes 1 and 4, homozygous (not informative); lanes 2 and 5, heterozygous (informative) without LOH; lanes 3 and 6, heterozygous with LOH; lane M, DNA marker pBR 322 DNA MspI digest. The gel is silver stained.
LOH at the nm23-H1 gene locus and clinical parameters
No correlation was found between nm23-H1 gene LOH and the age or sex of patients with informative tumour samples (table 3).
In addition, no correlation was found between nm23-H1 gene LOH and tumour size (p = 0.053; table 3).
A positive correlation was found between nm23-H1 gene LOH and the histological grade of tumours, although this was not significant because of the small sample size (table 3). Nm23-H1 gene LOH was found in three of 15 well differentiated, five of 18 moderately differentiated, and seven of 14 poorly differentiated tumours. A positive correlation was also found between nm23-H1 gene LOH and the Dukes’s stage of the tumour samples, although again this was not significant (table 3). Nm23-H1 gene LOH was found in three of 15 Dukes’s A, four of 16 Dukes’s B, and 12 of 37 Dukes’s C histologically staged tumours.
nm23-H1 mRNA expression and clinical parameters
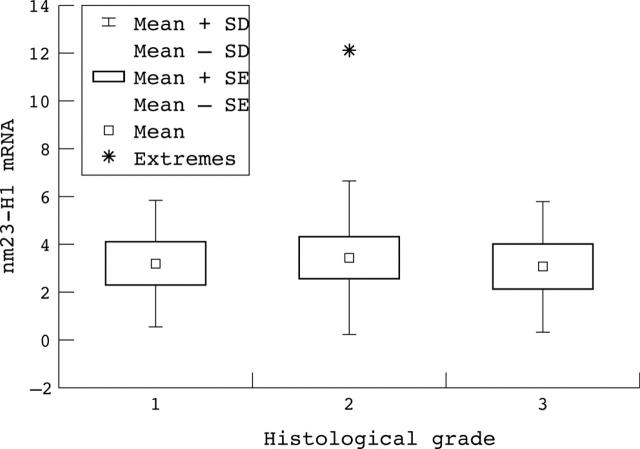
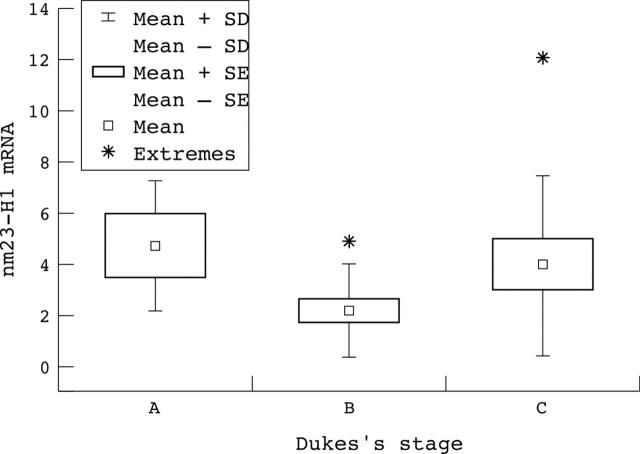
Thirty samples of colon cancer and corresponding normal tissue were examined for nm23-H1 mRNA expression. The expression of nm23-H1 mRNA was increased in 22 of these 30 colon tumours relative to that seen in the corresponding normal tissue. No significant correlation was found between nm23-H1 mRNA expression and histological grade (p = 0.957; fig 4), or Dukes’s stage (p = 0.159; fig 5) in the tumours examined.
Figure 4.
Correlation between nm23-H1 mRNA expression and the histological grade of the tumour.
Figure 5.
Correlation between nm23-H1 mRNA expression and the different Dukes’s stages of colon cancer.
DISCUSSION
In our study, we examined the expression of the nm23-H1 tumour suppressor protein in colorectal carcinomas and 41.2% of carcinomas showed weak expression of this protein. The remaining samples were negative for the nm23-H1 protein, as were all of the samples of normal colon and adenoma. The staining pattern was very similar in all carcinomas, namely: weak cytoplasmic staining of tumour cells with negative surrounding tissue. The nm23-H1 gene product has been identified as a potential metastasis suppressor, but its role in human cancers is still controversial.
The expression of the nm23-H1 protein has been examined in many different human tumours. It was found to be expressed in 18% of squamous cell lung carcinomas,24 22% of renal cell carcinomas,25 38% of oesophageal squamous cell carcinomas,26 84.5% of gastric carcinomas,27 66% of breast carcinomas,28 and 79% of colorectal carcinomas.29
Volm and co-workers detected a significant correlation between proliferation and nm23-H1 expression in non-small cell lung carcinomas. In addition, a direct correlation between apoptosis and nm23-H1 expression or between myc and nm23-H1 expression was found.30
Cytoplasmic nm23 protein concentrations are reduced in renal cell carcinoma, although the staining intensity shows no correlation with tumour stage, tumour grade, or prognosis. The reduction in nm23 protein concentrations may play a role during renal cell carcinoma pathogenesis, but not in progression or metastasis suppression.31
The results of Schneider et al indicate that nm23 overexpression is associated with a significantly worse prognosis in the early stage of well differentiated epithelial ovarian carcinoma.32 However, the prognostic relevance of nm23 expression in patients with advanced ovarian cancer was not confirmed.33
Mao and co-workers showed a negative correlation between nm23-H1 protein expression and tumour progression in human breast cancer. They also showed a significant difference (p < 0.001) in overall survival between patients with nm23-H1 negative and positive tumours.28
The overall survival rates of nm23-H1 negative patients with oesophageal squamous cell carcinoma were significantly shorter than those of nm23-H1 positive patients in the study of Iizuka and co-workers.34 In the study of Tomita et al, an inverse correlation was found between nm23-H1 protein expression and lymphatic vessel invasion in oesophageal squamous cell carcinoma. The overall survival rate was not different between nm23-H1 protein positive and negative patients. However, reduced expression of the nm23-H1 protein was associated with shorter overall survival in patients with involved lymph nodes.26 In a recent paper, Sarris and co-workers showed that an overall increase in nm23 expression or increase in nm23 expression in the cytoplasm of cells may be important in the early development of oesophageal adenocarcinoma, but that increased amounts of nuclear nm23 occur in the progression to metastatic disease.35
Müller and co-workers detected nm23-H1 expression in 84.5% of gastric cancer samples, and expression was positively correlated with the intestinal type of tumour, aggressive tumour growth, blood and lymphatic vessel invasion, and poor prognosis.27 The results of Nakamura et al suggest that the expression of nm23 in gastric cancer lesions correlates with tumour progression and/or proliferation rather than the suppression of metastasis.36 Nesi and co-workers showed that the expression of the nm23 gene in gastric carcinoma is significantly related to tumour progression and poor prognosis at five years.37
A significant association was found between nm23-H1 expression and the depth of invasion, lymph node involvement, and prognosis in human anal canal carcinoma. There was no significant association between nm23-H1 expression, histological type, and the age of the patients. Indinnimeo et al concluded that overexpression of the nm23-H1 protein in anal canal carcinoma may have implications for its metastatic potential.15
The expression of nm23-H1 in colon cancer has been examined by many groups of workers. Cheah et al showed that nm23-H1 expression inversely correlated with tumour staging. This result implies that nm23-H1 expression is raised in early stage colorectal carcinomas with a lower metastatic potential compared with late stage tumours with a higher metastatic potential. However, nm23-H1 protein expression was not significantly correlated with overall five year survival, disease recurrence, tumour differentiation, age, or sex.29 Our results are in agreement with their findings. In our study, no correlation was found between nm23-H1 protein positivity and the age of patients, although there was a significant difference between the male and the female groups of patients according to nm23-H1 protein positivity (p = 0.024). No correlation was found between nm23-H1 expression and size, histological grade, or Dukes’s stage of the tumours. However, most of the nm23-H1 positive tumours were non-metastatic stage, Dukes’s B tumours. This result is in agreement with the anti-metastatic role of this protein in human cancer. It might suppress the metastasis of these tumours staged as Dukes’ B in local lymph nodes or other organs. In our study, a negative correlation was found between nm23-H1 positivity and the histological grade of the colon adenocarcinomas. It is known that the histological grade is an independent prognostic factor in patients suffering from colon cancer. The correlation between high histological grade and low nm23-H1 protein positivity suggests that negative or low nm23-H1 protein expression could serve as a practical tumour marker. Tabuchi et al showed nm23-H1 expression in 44% of colorectal cancers, but an association with metastasis, histological stage, or survival was not found.38 Zeng and co-workers examined the expression of nm23-H1 in colorectal cancer and liver metastases, and correlated nm23-H1 expression with clinicopathological variables. No significant association was seen between the amount of nm23-H1 RNA and the patient’s age or sex, tumour location, differentiation, presence of lymph node involvement, or distant metastases. There appeared to be a trend between increasing relative nm23-H1 RNA and bowel wall invasion, irrespective of metastatic status.39
“Most of the nm23-H1 positive tumours in our study were non-metastatic stage, Dukes’s B tumours”
LOH at nm23-H1 was also examined in different human tumours and was found in 10% of squamous cell lung carcinomas,24 14% of renal cell carcinomas,40 22% of ovarian carcinomas,41 14% of gastric cancers,42 and 57% of colon cancers.43
Berney et al detected nm23-H1 LOH in 57% of sporadic colorectal cancers. nm23-H1 gene LOH was examined using four microsatellite loci spanning the 17q21–23 region and an increasing fraction of loci showing LOH was positively associated with the risk of liver metastasis.43 In our study nm23-H1 LOH was detected in 27.4% of sporadic colon cancers. No correlation was found between nm23-H1 LOH and tumour size. However, a positive correlation was found between nm23-H1 LOH and the histological grade of the tumours. A positive correlation was also found between nm23-H1 LOH and the Dukes’s stage of the tumour samples. Nm23-H1 gene LOH was most frequent in poorly differentiated tumours (50%) and Dukes’s C stage tumours. These two findings could be explained by the metastasis suppressor role of this gene in colon cancer.
Okada and co-workers reported that 29 of 42 colorectal carcinoma samples were informative and that LOH was found in three of these 29. In all three samples with LOH, cancer tissues expressed lower amounts of nm23-H1 mRNA when compared with those without LOH.44
In patients suffering from colorectal carcinoma with nm23-H1 LOH the risk of development of distant metastases was three times higher than in patients without nm23-H1 deletions in the five year follow up study of Cohn and colleagues.18
Heide et al examined the expression of nm23-H1 and nm23-H1 LOH in the liver metastases of colorectal cancer. Amounts of nm23-H1 mRNA in the metastatic tissues were not significantly different from those in normal colon mucosa and no LOH could be detected in the liver metastases.3
Take home messages.
Our results suggest that higher nm23-H1 protein expression in early stages of colon cancer may have a role in suppressing metastasis
In later stages both lower nm23-H1 protein expression and loss of heterozygosity of the nm23-H1 gene may play a role in colon cancer progression and metastasis
In our study, 30 samples of colon cancer and corresponding normal tissue were examined for nm23-H1 mRNA expression. Expression of nm23-H1 mRNA was increased in 22 of 30 colon tumours relative to that in corresponding normal tissue. Our results are in agreement with the results of Myeroff and Markowitz,19 who found increased expression of nm23-H1 mRNA in 80% of colon tumours relative to that in matched normal mucosa. In our study, no significant correlation was found between nm23-H1 mRNA expression and the histological grade or Dukes’s stage of the tumours.
In summary, our findings suggest that higher nm23-H1 protein expression in early stages may have a role in suppressing metastasis in sporadic colon cancer, whereas at a later stage both lower nm23-H1 protein expression and LOH of the nm23-H1 gene may play a role in colon cancer progression and metastasis.
Abbreviations
LOH, loss of heterozygosity
NDP, nucleoside diphosphate
PBS, phosphate buffered saline
PCR, polymerase chain reaction
RT, reverse transcription
VNTR, variable nucleotide tandem repeat
REFERENCES
- 1.Wang L, Patel U, Ghosh L, et al. Mutation in the nm23 gene is associated with metastasis in colorectal cancer. Cancer Res 1993;53:717–20. [PubMed] [Google Scholar]
- 2.MacDonald NJ, De la Rosa A, Steeg PS. The potential roles of nm23 in cancer metastasis and cellular differentiation. Eur J Cancer 1995;31A:1096–100. [DOI] [PubMed] [Google Scholar]
- 3.Heide I, Thiede C, Poppe K, et al. Expression and mutational analysis of nm23-H1 in liver metastases of colorectal cancer. Br J Cancer 1994;70:1267–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steeg PS, Bevilacqua G, Kopper L, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst 1988;80:200–4. [DOI] [PubMed] [Google Scholar]
- 5.Stahl JA, Leone A, Rosengard AM, et al. Identification of a second human nm23 gene, nm23-H2. Cancer Res 1991;51:445–9. [PubMed] [Google Scholar]
- 6.MacDonald NJ, De la Rosa A, Benedict MA, et al. A serine phosphorylation of nm23, and not its nucleoside diphosphate kinase activity, correlates with suppression of tumor metastatic potential. J Biol Chem 1993;268:25780–9. [PubMed] [Google Scholar]
- 7.Freije JM, Blay P, MacDonald NJ, et al. Site-directed mutation of nm23-H1. Mutations lacking motility suppressive capacity upon transfection are deficient in histidine-dependent protein phosphotransferase pathways in vitro. J Biol Chem 1997;272:5525–32. [DOI] [PubMed] [Google Scholar]
- 8.De la Rosa A, Williams RL, Steeg PS. Nm23/nucleoside diphosphate kinase: toward a structural and biochemical understanding of its biological functions. Bioessays 1995;17:53–62. [DOI] [PubMed] [Google Scholar]
- 9.Pavelić K, Kapitanović S, Radošević S, et al. Increased activity of nm23-H1 gene in squamous cell carcinoma of the head and neck is associated with advanced disease and poor prognosis. J Mol Med 2000;78:111–18. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi A, Urana T, Fushida S, et al. Inverse association of nm23-H1 expression by colorectal cancer with liver metastasis. Cancer 1993;68:1020–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez JA, Prevot S, Nordlinger B, et al. Overexpression of nm23-H1 and nm23-H2 genes in colorectal carcinomas and loss of nm23-H1 expression in advanced tumour stages. Gut 1995;37:712–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tannapfel A, Kockerling F, Katalinic A, et al. Expression of nm23-H1 predicts lymph node involvement in colorectal carcinoma. Dis Colon Rectum 1995;38:651–4. [DOI] [PubMed] [Google Scholar]
- 13.Royds JA, Cross SS, Silcocks PB, et al. Nm23 “anti-metastatic” gene product expression in colorectal carcinoma. J Pathol 1994;172:261–6. [DOI] [PubMed] [Google Scholar]
- 14.Lindmark G. Nm23-H1 immunohistochemistry is not useful as predictor of metastatic potential of colorectal cancer. Br J Cancer 1996;74:1413–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Indinnimeo M, Cicchini C, Stazi A, et al. Correlation between nm23-H1 overexpression and clinicopathological variables in human anal canal carcinoma. Oncol Rep 1999;6:1353–6. [DOI] [PubMed] [Google Scholar]
- 16.Cohn KH, Wang FS, Desoto-LaPaix F, et al. Association of nm23-H1 allelic deletions with distant metastases in colorectal carcinoma. Lancet 1991;338:722–4. [DOI] [PubMed] [Google Scholar]
- 17.Leone A, McBride OW, Weston A, et al. Somatic allelic deletion of nm23 in human cancer. Cancer Res 1991;51:2490–3. [PubMed] [Google Scholar]
- 18.Cohn KH, Ornstein DL, Wang F, et al. The significance of allelic deletions and aneuploidy in colorectal carcinoma. Cancer 1997;79:233–44. [PubMed] [Google Scholar]
- 19.Myeroff LL, Markowitz SD. Increased nm23-H1 and nm23-H2 messenger RNA expression and absence of mutations in colon carcinomas of low and high metastatic potential. J Natl Cancer Inst 1993;85:147–52. [DOI] [PubMed] [Google Scholar]
- 20.Cawkwell L, Lewis FA, Quirke P. Frequency of allele loss of DCC, p53, RB1, WT1, NF1, NM23, APC/MCC in colorectal cancer assayed by fluorescent multiplex polymerase chain reaction. Br J Cancer 1994;70:813–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spaventi R, Pečur L, Pavelić K, et al. Human tumor bank in Croatia: a possible model for a small bank as a part of the future European tumor bank network. Eur J Cancer 1994;30A:419. [DOI] [PubMed] [Google Scholar]
- 22.Turnubull RB, Kyle K, Watson FR, et al. Cancer of the rectum: the influence of the no-touch isolation technique on survival rates. Ann Surg 1967;166:420–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deans GT, Parks TG, Rowlands BJ, et al. Prognostic factors in colorectal cancer. Br J Surg 1992;79:608–13. [DOI] [PubMed] [Google Scholar]
- 24.Herak Bosnar M, Pavelić K, Hrašćan R, et al. Loss of heterozygosity of the nm23-H1 gene in human renal cell carcinomas. J Cancer Res Clin Oncol 1997;123:485–8. [DOI] [PubMed] [Google Scholar]
- 25.Ljungberg B, Osterdahl B, Mehle C. Clinical significance of nm23 expression in renal cell carcinoma. Urol Res 1999;27:103–7. [DOI] [PubMed] [Google Scholar]
- 26.Tomita M, Ayabe T, Matsuzaki Y, et al. Expression of nm23-H1 gene product in esophageal squamous cell carcinoma and its association with vessel invasion and survival. BMC Cancer 2001;1:3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller W, Schneiders A, Hommel G, et al. Expression of nm23 in gastric carcinoma. Cancer 1998;83:2481–7. [DOI] [PubMed] [Google Scholar]
- 28.Mao H, Liu H, Fu X, et al. Loss of nm23 expression predicts distal metastases and poorer survival for breast cancer. Int J Oncol 2001;18:587–91. [DOI] [PubMed] [Google Scholar]
- 29.Cheah PY, Cao X, Eu KW, et al. Nm23-H1 immunostaining is inversely associated with tumour staging but not overall survival or disease recurrence in colorectal carcinomas. Br J Cancer 1998;77:1164–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volm M, Mattern J, Koomagi R. Association between nm23-H1 expression, proliferation and apoptosis in non-small cell lung carcinomas. Clin Exp Metastasis 1998;16:595–602. [DOI] [PubMed] [Google Scholar]
- 31.Ayhan A, Usubutun A, Ozen H, et al. nm23 protein expression in renal cell tumors: the role of the cell type. Oncol Rep 1998;5:979–83. [DOI] [PubMed] [Google Scholar]
- 32.Schneider J, Pollan M, Jimenez E, et al. nm23-H1 expression defines a high-risk subpopulation of patients with early-stage epithelial ovarian carcinoma. Br J Cancer 2000;82:1662–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baekelandt M, Holm R, Trope CG, et al. The significance of metastasis-related factors cathepsin-D and nm23 in advanced ovarian cancer. Ann Oncol 1999;10:1335–41. [DOI] [PubMed] [Google Scholar]
- 34.Iizuka N, Tangoku A, Hayashi H, et al. The association between nm23-H1 expression and survival in patients with esophageal squamous cell carcinoma. Cancer Lett 1999;138:139–44. [DOI] [PubMed] [Google Scholar]
- 35.Sarris M, Konopka M, Lee CS. Differential expression of the nm23 protein in the progression of oesophageal adenocarcinoma. Pathology 2003;35:37–41. [PubMed] [Google Scholar]
- 36.Nakamura T, Tabuchi Y, Ohno M. Relations of nm23 expression to clinicopathologic variables and proliferative activity of gastric cancer lesions. Cancer Detect Prev 1998;22:246–50. [DOI] [PubMed] [Google Scholar]
- 37.Nesi G, Palli D, Pernice LM, et al. Expression of nm23 gene in gastric cancer is associated with a poor 5-year survival. Anticancer Res 2001;21:3643–9. [PubMed] [Google Scholar]
- 38.Tabuchi Y, Nakamura T, Kuniyasu T, et al. Expression of nm23-H1 in colorectal cancer: no association with metastases, histological stage, or survival. Jpn J Surg 1999;29:116–20. [DOI] [PubMed] [Google Scholar]
- 39.Zeng ZS, Hsu S, Zhang ZF, et al. High level of nm23-H1 gene expression is associated with local colorectal cancer progression not with metastases. Br J Cancer 1994;70:1025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herak Bosnar M, Pavelić K, Križanac Š, et al. Squamous cell lung carcinomas: the role of nm23-H1 gene. J Mol Med 1997;75:609–13. [DOI] [PubMed] [Google Scholar]
- 41.Mandai M, Konishi I, Komatsu T, et al. Mutation of the nm23 gene, loss of heterozygosity at the nm23 locus and K-ras mutation in ovarian carcinoma: correlation with tumor progression and nm23 gene expression. Br J Cancer 1995;72:691–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang BG, Park IC, Park MJ, et al. Role of the nm23-H1 gene in the metastasis of gastric cancer. J Korean Med Sci 1997;12:514–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berney CR, Fisher RJ, Yang JI, et al. Genomic alterations (LOH, MI) on chromosome 17q21–23 and prognosis of sporadic colorectal cancer. Int J Cancer 2000;89:1–7. [DOI] [PubMed] [Google Scholar]
- 44.Okada K, Urano T, Goi T, et al. Isolation of human nm23 genomes and analysis of loss of heterozygosity in primary colorectal carcinomas using a specific genomic probe. Cancer Res 1994;54:3979–82. [PubMed] [Google Scholar]