Abstract
Background: Loss of imprinting (LOI) of the H19/IGF2 domain is a common feature of Wilms tumour. The GTL2/DLK1 domain is also imprinted and is structurally similar to H19/IGF2. The question arises as to whether DLK1 also undergoes LOI in Wilms tumour, or whether the LOI mechanism is restricted to the H19/IGF2 domain.
Aim: To investigate the imprinting status of DLK1 in Wilms tumours with IGF2 LOI. The cellular localisation of DLK1 in the tumours was also examined.
Methods: DLK1 expression was measured by quantitative real time polymerase chain reaction (Q-PCR) in 30 Wilms tumours that had previously been classified according to whether they had IGF2 LOI, WT1 mutations, or 11p15.5 loss of heterozygosity. Allele specific expression of DLK1 was examined by direct sequencing using a DLK1 exon 5 polymorphism (rs1802710). Immunohistochemical analysis of DLK1 was performed on 13 tumours and two intralobar nephrogenic rests, in addition to two fetal kidneys and one fetal skeletal muscle sample.
Results: Ten of 30 tumours were heterozygous for rs1802710 and all tumours showed retention of imprinting of DLK1. Moderate to high expression of DLK1 was detected by Q-PCR in nine of 13 tumours with myogenic differentiation. Immunohistochemical expression of DLK1 was detected in the myogenic elements.
Conclusion: LOI does not occur at the GTL2/DLK1 domain in Wilms tumour. This finding suggests that LOI at 11p15.5 does not reflect non-specific disruption of a shared imprinting mechanism. DLK1 expression in Wilms tumour might reflect the presence of myogenic differentiation, rather than an alteration of its imprinting status.
Genomic imprinting is a mechanism by which there is preferential expression of an allele depending on the parental origin. Two well known reciprocally imprinted genes, IGF2 and H19, are located on human chromosome 11p15.5.1 IGF2 is exclusively expressed from the paternal allele, whereas the H19 allele is silenced on that allele.1 The transcription of these genes is regulated by a differentially methylated region (DMR) upstream of the H19 promoter and by an H19 downstream enhancer (fig 1).2–5 The DMR contains binding sites for CTCF, a chromatin insulator4,5 that binds to the unmethylated DMR on the maternal allele, preventing IGF2 transcription and enabling H19 transcription. On the paternal allele, methylation of the DMR prevents CTCF binding, leading to IGF2 transcription and H19 silencing.4,5
Figure 1.
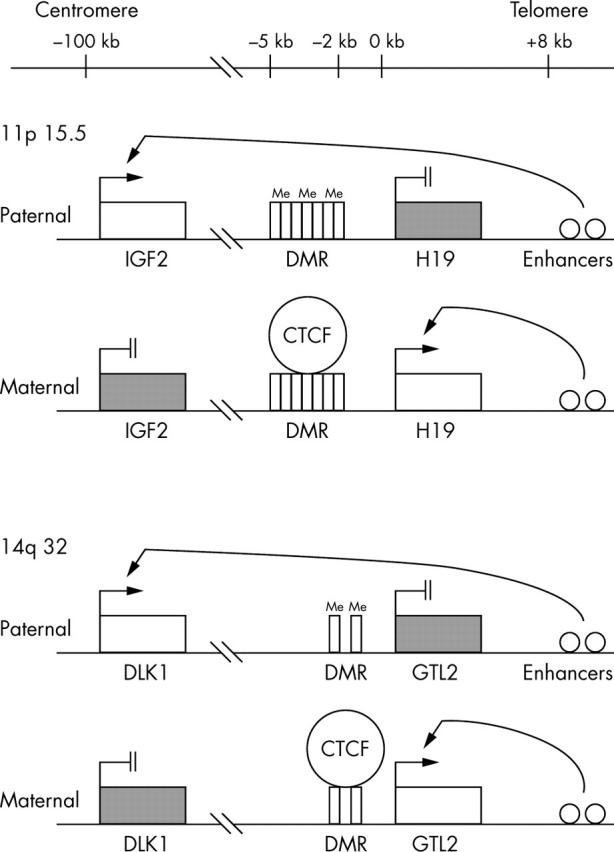
The structural, spatial, and regulatory features of IGF2/H19 and DLK1/GTL2 domains. Expressed and imprinted genes are shown as open boxes and black boxes, respectively. CTCF binding sites are indicated by vertical rectangles. Open circles indicate enhancer elements.
“Loss of imprinting is an epigenetic error associated with tumorigenesis, particularly in Wilms tumour”
An imprinted domain (DLK1/GTL2) has recently been reported on human chromosome 14q32.6,7 DLK1 (δ-like, homologue-drosophila) is a paternally expressed gene6,7 that is homologous to the Notch/Delta family of developmentally regulated signalling molecules.8–10 DLK1 is thought to play a role in the development and differentiation of the adrenal medulla, adipocytes, haemopoietic progenitor cells, and osteoblasts.11–14 GTL2 is an untranslated maternally expressed gene.15 The DLK1/GTL2 domain has striking similarities to the IGF2/H19 domain (fig 1).6 The similarities are as follows:
The maternally expressed, non-coding GTL2 and H19 transcripts are both separated by approximately 100 kb from their corresponding paternally expressed, protein encoding genes, DLK1 and IGF2.6,15
Similar to IGF2 and H19, DLK1 and GTL2 are transcribed in the same orientation and are reciprocally imprinted.6,15
Both H19 and GTL2 have DMRs containing CTCF binding sites for the formation of a chromatin boundary.6,15
Similar enhancer consensus sequences are located approximately 8 kb downstream from the H19 and GTL2 transcription start sites.2,6
Loss of imprinting (LOI) is an epigenetic error associated with tumorigenesis, particularly in Wilms tumour.16,17 The aim of our study was to investigate whether tumours with IGF2 LOI also have LOI of DLK1, or whether the LOI mechanism is restricted to the H19/IGF2 domain. Imprinting status, expression, and cellular localisation of DLK1 were examined in 30 Wilms tumours. We also investigated whether the expression of DLK1 in Wilms tumours is associated with rhabdomyogenic differentiation because mutations in a regulatory locus of DLK1 lead to muscle hypertrophy in callipyge sheep.18
MATERIAL AND METHODS
Sample collection and histology
Thirty Wilms tumours used for our study were selected from a collection of tumours obtained with informed consent from North Health ethics committee, Auckland, New Zealand. From this collection, all tumours with IGF2 LOI and WT1 mutations (17 and eight, respectively) were included in this study, and five tumours with 11p15.5 loss of heterozygosity (LOH) were chosen at random. IGF2 LOI and WT1 mutations were measured as described previously.17,19 LOH at 11p15.5 was assessed either by Southern blotting with Ha-Ras VNTR (variable number of random repeats), INS VNTR, and globin probes or by DNA microsatellite analysis with tyrosine hydroxylase.20 The tumours were examined histologically for the presence of myogenic differentiation; that is, the presence of muscle cells with striation (rhabdomyogenesis). Thirteen of the 30 tumours and two intralobar nephrogenic rest (ILNR) lesions were also available for immunohistochemical study. Fetal kidneys (8 and 21 weeks) and fetal skeletal muscle samples (8 weeks) were also used for immunohistochemistry.
Analysis of heterozygosity for DLK1
Wilms tumours were examined for a polymorphism at exon 5 (rs1802710) in the DLK1 gene by direct sequencing using primers DLK-F9 and DLK-R7 (see below). Analysis of DLK1 imprinting was performed for the informative cases. To confirm DNA heterozygosity and allele specific expression, parental DNA was also analysed.
Analysis of DLK1 imprinting
Total RNA (2 μg) was DNase treated and cDNA was synthesised using Superscript TMII RNase H− reverse transcriptase according to the manufacturer’s protocol (Life Technologies, Gaithersburg, Maryland, USA). To eliminate the possibility of contamination by genomic DNA, cDNA was amplified using primers (F9 and R7) spanning exons 4 and 5. DLK1 genomic DNA and cDNA were amplified (35 cycles at 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds) and PCR products were purified using a Qiagen kit (Qiagen, Hilden, Germany). Sequence reactions were performed with the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, California, USA) and run on an ABI 3100 sequencer. To confirm the sequencing results, all samples were sequenced from both directions using the forward and reverse primers (F9 and R7). The primers have been reported previously6 (DLK-F9, AAC AAC GGG ACC TGC GTG; DLK-R7, GCT TGC ACA GAC ACT CGT AGC TCA CC).
Quantitative real time PCR
Quantitative real time PCR (Q-PCR) was performed with an ABI PRISM 7900HT Sequence Detection System using ABI TaqMan probes, primers for DLK1 (Hs00171584, ABI), and Absolute QPCR ROX mix. The expression of DLK1 in each tumour sample was normalised to an endogenous control gene UBE2G2 (ubiquitin conjugating enzyme E2G2) and expressed relative to a pooled reference RNA sample.
Immunohistochemistry
Paraffin wax embedded sections (4 μm thick) were treated with 3% hydrogen peroxide for 15 minutes. Slides were autoclaved at 121°C for 15 minutes in 10mM citrate buffer. Sections were incubated for 30 minutes in 5% normal rabbit serum in phosphate buffered saline (PBS) with 0.1% bovine serum albumin (BSA). The sections were incubated with anti-DLK1 antibody (SC-8623; Santa Cruz Biotechnology, California, USA; 1/20 dilution) in PBS containing BSA at room temperature for one hour, followed by overnight incubation at 4°C. As a negative control, adjacent sections were incubated without primary antibody. Tissue sections were washed in PBS and reincubated for 30 minutes with horseradish peroxidase congugated rabbit antigoat IgG (H+L) (Zymed, South San Francisco, California, USA), diluted 1/200 in PBS with BSA. Sections were incubated with diaminobenzidine substrate (Rockland, Gilbertsville, Pennsylvania, USA) and counterstained with haematoxylin.
RESULTS
Expression of DLK1 RNA and histological correlation in Wilms tumour
DLK1 RNA expression was examined by Q-PCR (table 1). The highest expression levels were detected in the tumours with myogenic differentiation (rhabdomyogenesis); that is, five WT1 mutant tumours (WT1-1, WT1-2, WT1-3, WT1-5, and WT1-8) and two IGF2 LOI tumours with myogenic differentiation (LOI-15 and LOI-16). Although all eight WT1 mutant tumours had rhabdomyogenesis, three tumours (WT1-4, WT1-6, and WT1-7) showed low DLK1 expression. Most tumours with IGF2 LOI were composed predominantly of blastemal cells and showed low expression of DLK1, whereas two tumours without myogenic differentiation (LOI-9 and LOI-11, both of which showed mixed epithelial and blastemal histology) had moderate to high expression of DLK1. The tumour (LOI-11) with the highest DLK1 expression had blastemal predominant histology with focal anaplasia, and contained a large area of necrosis surrounded by granulation tissue with aggregated foamy macrophages. Moderate DLK1 expression was detected in three tumours with 11p15.5 LOH (LOH-1, LOH-2, and LOH-3). One of these tumours (LOH-1) contained myogenic differentiation, but the remaining two tumours showed blastemal predominant histology without myogenic differentiation.
Table 1.
Expression and imprinting of DLK1, and histology of 30 Wilms tumours
| DLK1 gene | Histology | Myogenesis | |||
| Q-PCR | Genotype | Allelic expression | |||
| IGF2 LOI tumours | |||||
| LOI-1 | 0.02 | T/C | C/– | Epi | – |
| LOI-2 | 0.1 | T/C | T/– | Bl | – |
| LOI-3 | 0.1 | T/C | C/– | Bl | – |
| LOI-4 | 0.1 | T/C | T/– | Bl | – |
| LOI-5 | NA | T/C | C/– | Tri | ++ |
| LOI-6 | 0.9 | T | NI | Bl | – |
| LOI-7 | 0.1 | T | NI | Bl | – |
| LOI-8 | NA | C | NI | Bl | – |
| LOI-9 | 8.2 | C | NI | Bl/Epi | – |
| LOI-10 | 0.2 | C | NI | Bl | – |
| LOI-11 | 41.6 | C | NI | Epi/Bl | – |
| LOI-12 | 0.4 | T | NI | Bl | – |
| LOI-13 | 0.5 | C | NI | Tri | + |
| LOI-14 | 0.1 | T | NI | Bl | – |
| LOI-15 | 5.6 | C | NI | Tri | ++ |
| LOI-16 | 2.3 | C | NI | Tri | ++ |
| LOI-17 | NA | T | NI | Bl | – |
| WT1 mutant tumours | |||||
| WT1–1 | 4.5 | T/C | T/– | Str | ++ |
| WT1–2 | 15.2 | T/C | T/– | Str | ++ |
| WT1–3 | 9.7 | T/C | C/– | Str | ++ |
| WT1–4 | 0.7 | T | NI | Str | ++ |
| WT1–5 | 14 | C | NI | Str | +++ |
| WT1–6 | 0.4 | C | NI | Str | ++ |
| WT1–7 | 0.4 | C | NI | Str | ++ |
| WT1–8 | 12.7 | T | NI | Str | +++ |
| 11p15.5 LOH tumours | |||||
| LOH-1 | 2.9 | T/C | T/– | Tri | ++ |
| LOH-2 | 2.2 | T/C | T/– | Bl | – |
| LOH-3 | 6.7 | C | NI | Bl/Epi | – |
| LOH-4 | 0.1 | T | NI | Bl | – |
| LOH-5 | 0.5 | C | NI | Bl | – |
Bl, blastemal predominant; Epi, epithelial predominant; LOH, loss of heterozygosity; LOI, loss of imprinting; NA, not available; NI, not informative; Q-PCR, quantitative real time polymerase chain reaction; Str, stromal predominant; Tri, triphasic; absent –, low +, moderate ++, high +++.
Imprinting status of DLK1 in Wilms tumours
Ten of the 30 tumours were heterozygous for the T/C polymorphism at exon 5 (rs1802710), namely: five tumours with IGF2 LOI, three tumours with WT1 mutations, and two tumours with 11p15.5 LOH. Imprinting (monoallelic expression) of DLK1 was seen in all heterozygous tumours, irrespective of the imprinting status of IGF2 and/or level of DLK1 expression. Heterozygosity and parental specific expression of DLK1 in the tumours were confirmed by analysing the parental DNA (fig 2).
Figure 2.
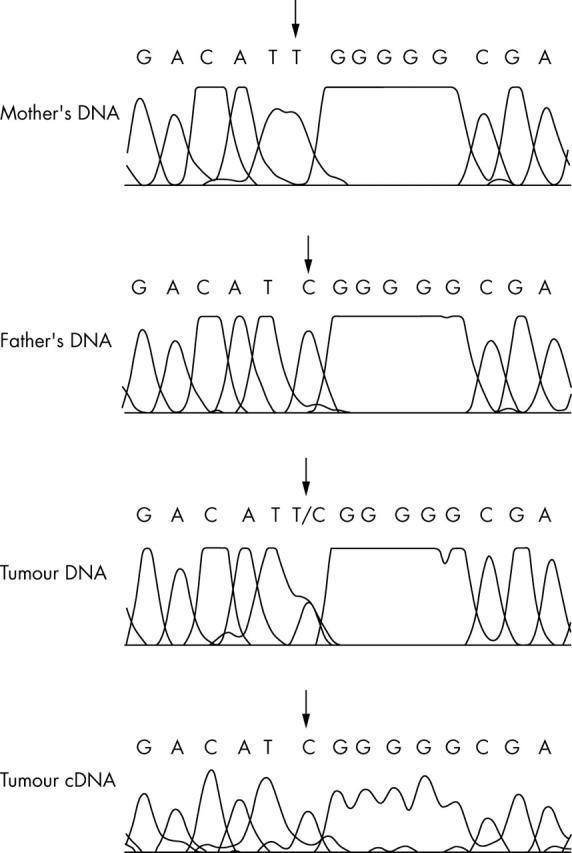
The tumour DNA is heterozygous for the C/T polymorphism. The father and mother are homozygous for the C and T allele, respectively. Therefore, exclusive expression of the C allele indicates paternal expression of DLK1.
Cellular localisation of DLK1
The localisation of DLK1 was first examined in the developing kidney because Wilms tumour often recapitulates early kidney epithelial development. Expression of DLK1 is probably required for the later stages of epithelial kidney development rather than the early stages, because DLK1 immunostaining was faintly positive or negative in the uninduced mesenchyme, condensed mesenchyme, renal vesicles, comma shaped bodies, S shaped bodies, presumptive podocytes, and glomerular epithelia (fig 3A), and weakly positive in the cytoplasm of ureteric buds and ureteric bud ampullae (fig 3A). DLK1 was moderately to highly expressed in the cytoplasm of developing tubules (future proximal and distal convoluted tubules) (fig 3A). DLK1 was also expressed in the apical membrane of distal tubules or Henle’s loop and localised in the cytoplasm and cell membrane in collecting ducts. Three normal kidney tissues adjacent to the tumours (WT1-1, WT1-2, and WT1-3) were available for immunohistochemistry. Moderate to strong expression of DLK1 was seen in the nucleus and cytoplasm of collecting ducts, Henle’s loops, proximal and distal tubules (fig 3B). Weak expression of DLK1 was sporadically seen in the nuclei of glomerular capillary endothelia in both immature and mature glomeruli. DLK1 was also weakly expressed in the smooth muscles of small arteries and nuclei in endothelia. Skeletal muscles in a fetus (8 weeks) showed DLK1 expression (fig 3C), indicating that DLK1 is associated with myogenic differentiation.
Figure 3.
Immunohistochemistry for DLK1 in (A) the developing kidney (21 weeks), (B) tumour bearing normal kidney, and (C) fetal skeletal muscle (8 weeks). Original magnification: A, ×200; B, C, ×400. (A) DLK1 immunostaining is faintly positive or negative in condensed mesenchyme (cm), renal vesicles (rv), an S shaped body (s), a presumptive podocyte (pp), and glomerular epithelium (gl). DLK1 was weakly positive in the cytoplasm of ureteric buds (ub) and ureteric bud ampullae (uba). DLK1 was moderately to highly expressed in the cytoplasm of developing tubules (dt, future proximal and distal convoluted tubules). (B) Expression of DLK1 was seen in the nucleus and cytoplasm of collecting ducts (cd), Henle’s loops (hl), proximal and distal convoluted tubules (pct and dct). (C) DLK1 is expressed in fetal skeletal muscle.
Thirteen Wilms tumours were analysed for DLK1 using immunohistochemistry (table 2): three IGF2 LOI tumours, eight WT1 mutant tumours, and two 11p15.5 LOH tumours that had detectable DLK1 expression by Q-PCR. Moderate to strong expression of DLK1 was seen in the cytoplasm and cell membrane of the myogenic elements in all tumours (fig 4A), including three WT1 mutant tumours that showed low expression of DLK1 in the Q-PCR assay. Weak to moderate expression of DLK1 was localised to the cytoplasm and nucleus of the epithelial components (fig 4A, B). Immunostaining for DLK1 was occasionally seen in the nucleus and cytoplasm of the blastemal component in all three molecular classes (fig 4A, B). The tumour with the highest DLK1 expression by Q-PCR (LOI-11) showed strong expression of DLK1 in the cytoplasm of foamy macrophages, suggesting that the high number of macrophages may have contributed to the high expression of DLK1 RNA in this tumour. The expression pattern of DLK1 in ILNRs (WT1-1 and WT1-2) was similar to that of Wilms tumours in that they had focal expression of DLK1 in the blastemal and epithelial components (fig 4C). Although myogenic elements were not seen in ILNRs, the non-polarised epithelial structures, sometimes referred to as “central and stromal tubules” that correspond to the collecting duct system,21 showed high DLK1 staining, similar to that seen in normal kidney.
Table 2.
Immunohistochemical localisation and staining intensity of DLK1 in Wilms tumours
| Tumour group | Tumour no | Epi | Bl | Str |
| LOI tumours with myogenesis | LOI-15 and LOI-16 | −/+ | −/+ | +/++ (myogenic) |
| LOI tumours without myogenesis | LOI-11 | −/+ | – | +++ (macrophage) |
| WT1 mutant tumours | WT1-1 to WT1-8 | −/+ | −/+ | ++/+++ (myogenic) |
| ILNRs | WT1-1 and WT1-2 | +/++ | −/+ | – |
| LOH tumours | LOH-2 and LOH-3 | −/+ | −/+ | – |
Each column shows the range of expression: no detectable expression, –; mild, +; moderate, ++; high expression, +++.
Bl, blastemal; Epi, epithelial; ILNRs, intralobar nephrogenic rests; LOH, loss of heterozygosity; LOI, loss of imprinting; Str, stromal.
Figure 4.
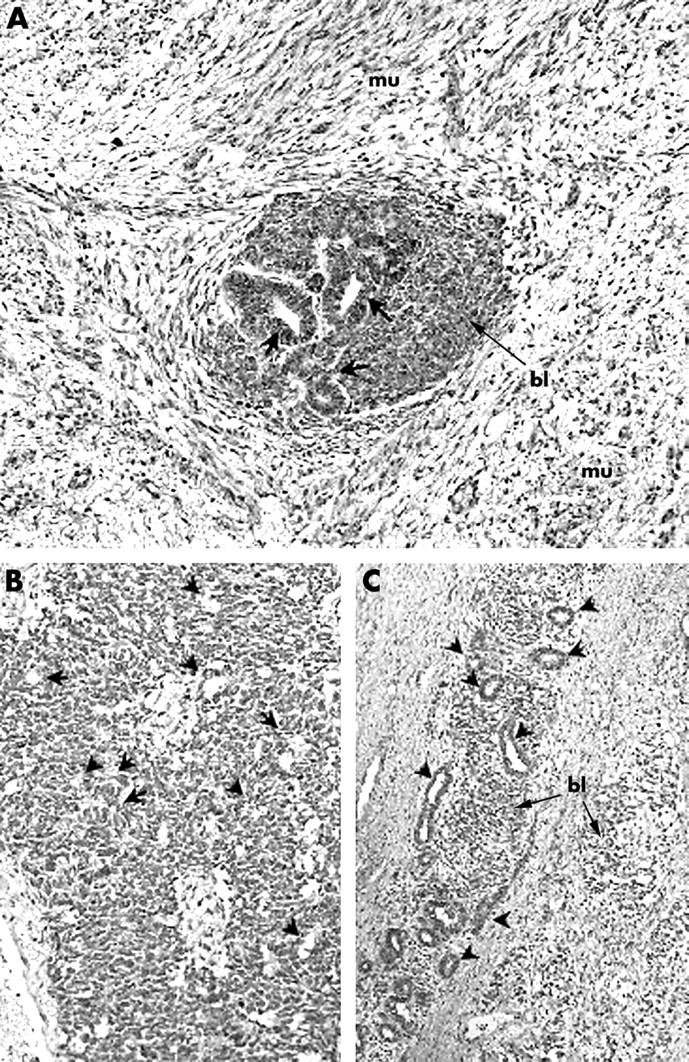
Immunohistochemistry for DLK1 in (A) Wilms tumour with myogenic differentiation, (B) Wilms tumour without myogenic differentiation, and (C) an intralobar nephrogenic rest (ILNR). Original magnification, ×200. (A) Tumour WT1-3. Immunostaining for DLK1 is seen in the cytoplasm of the myogenic elements (mu). Focal immunostaining is seen in the nucleus and cytoplasm of the blastemal (bl) and the epithelial cells (arrows). (B) Tumour LOH-3. A tumour showing blastemal predominant histology with focal epithelial differentiation (arrowheads). DLK1 is expressed in the nucleus and cytoplasm of the blastemal cells and epithelial elements. (C) An ILNR showing similar DLK1 staining patterns to that seen in Wilms tumours. Non-polarised epithelial structures (central and stromal tubules, indicated by arrowheads) and blastemal components (bl) show high expression of DLK1.
DISCUSSION
We have shown that LOI in Wilms tumour is restricted to the IGF2/H19 domain, even though the DLK1/GTL2 domain shares structural and epigenetic similarities with the IGF2/H19 domain. This finding suggests that IGF2 LOI does not reflect non-specific disruption of a shared imprinting mechanism. DLK1 has been shown to be expressed in neuroendocrine cancers, such as neuroblastoma, phaeochromocytoma, small cell carcinoma of the lung, and pancreatic islet tumours.8,22 Analysis of DLK1 imprinting has recently been reported in neuroblastoma, lymphoma, and brain tumours.13,23 However, none of these tumours showed loss of imprinting of DLK1.
“DLK1 might provide a useful marker for other tumours that show rhabdomyogenesis, such as rhabdomyosarcoma and pleuropulmonary blastoma”
Expression of DLK1 is associated with muscular growth and development, because DLK1 is highly expressed without alteration of its imprinting status in the callipyge phenotype in sheep that have an inherited skeletal muscular hypertrophy.18 Overexpression of DLK1 in the callipyge phenotype is seen preferentially in skeletal muscle,18 although it has been shown to be expressed in uterine leiomyomata,24 which are derived from smooth muscle cells. In our immunohistochemical study, expression of DLK1 was strong in fetal skeletal muscles, but weak in the smooth muscles of small arteries of tumour bearing kidneys. Similar to the observation of DLK1 overexpression in the callipyge phenotype in sheep, high amounts of DLK1 mRNA were found predominantly in Wilms tumours with abundant rhabdomyogenesis, many of which carried WT1 mutations. The localisation of DLK1 in the myogenic elements was confirmed by immunohistochemistry. The expression of DLK1 did not mimic that seen in early kidney epithelial development because expression of DLK1 was weak or absent in the developing kidney, whereas it was variable in the blastemal and epithelial components.
Take home messages.
Although loss of imprinting (LOI) of the H19/IGF2 domain is a common feature of Wilms tumour, we found no evidence of LOI of the structurally similar GTL2/DLK1 domain
This suggests that LOI at 11p15.5 does not reflect non-specific disruption of a shared imprinting mechanism, and that future studies into the mechanism of abnormal imprinting in Wilms tumour should focus on the specific genetic and epigenetic changes within the H19/IGF2 domain
DLK1 expression in Wilms tumour might reflect the presence of myogenic differentiation, rather than an alteration of its imprinting status
Our immunohistochemical study suggests that DLK1 might provide a useful marker for other tumours that show rhabdomyogenesis, such as rhabdomyosarcoma and pleuropulmonary blastoma. The specificity of loss of imprinting within the H19/IGF2 domain suggests that future studies into the mechanism of abnormal imprinting in Wilms tumour should focus on the specific genetic and epigenetic changes within the H19/IGF2 domain, rather than the abnormal function of a generalised imprinting mechanism.
Acknowledgments
We thank Dr M Sano, Department of Pathology, Nihon University School of Medicine, Japan, for helpful advice on immunohistochemistry. This work was supported by the Health Research Council of New Zealand, the Cancer Society of New Zealand, and the NZ Lottery Health Grants Board.
Abbreviations
BSA, bovine serum albumin
DMR, differentially methylated region
ILNR, intralobar nephrogenic rest
LOH, loss of heterozygosity
LOI, loss of imprinting
PBS, phosphate buffered saline
PCR, polymerase chain reaction
Q-PCR, quantitative real time polymerase chain reaction
REFERENCES
- 1.Zemel S, Bartolomei MS, Tilghman SM. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat Genet 1992;2:61–5. [DOI] [PubMed] [Google Scholar]
- 2.Leighton PA, Saam JR, Ingram RS, et al. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev 1995;9:2079–89. [DOI] [PubMed] [Google Scholar]
- 3.Frevel MA, Sowerby SJ, Petersen GB, et al. Methylation sequencing analysis refines the region of H19 epimutation in Wilms tumor. J Biol Chem 1999;274:29331–40. [DOI] [PubMed] [Google Scholar]
- 4.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 2000;405:482–5. [DOI] [PubMed] [Google Scholar]
- 5.Hark AT, Schoenherr CJ, Katz DJ, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 2000;405:486–9. [DOI] [PubMed] [Google Scholar]
- 6.Wylie AA, Murphy SK, Orton TC, et al. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res 2000;10:1711–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takada S, Paulsen M, Tevendale M, et al. Epigenetic analysis of the Dlk1–Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2–H19. Hum Mol Genet 2002;11:77–86. [DOI] [PubMed] [Google Scholar]
- 8.Laborda J, Sausville EA, Hoffman T, et al. dlk, a putative mammalian homeotic gene differentially expressed in small cell lung carcinoma and neuroendocrine tumor cell line. J Biol Chem 1993;268:3817–20. [PubMed] [Google Scholar]
- 9.Jensen CH, Krogh TN, Hojrup P, et al. Protein structure of fetal antigen 1 (FA1). A novel circulating human epidermal-growth-factor-like protein expressed in neuroendocrine tumors and its relation to the gene products of dlk and pG2. Eur J Biochem 1994;225:83–92. [DOI] [PubMed] [Google Scholar]
- 10.Lindsell CE, Boulter J, diSibio G, et al. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand–receptor pairs that may function in neural development. Mol Cell Neurosci 1996;8:14–27. [DOI] [PubMed] [Google Scholar]
- 11.Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 1993;73:725–34. [DOI] [PubMed] [Google Scholar]
- 12.Ohno N, Izawa A, Hattori M, et al. dlk inhibits stem cell factor-induced colony formation of murine hematopoietic progenitors: Hes-1-independent effect. Stem Cells 2001;19:71–9. [DOI] [PubMed] [Google Scholar]
- 13.Van Limpt VA, Chan AJ, Van Sluis PG, et al. High delta-like 1 expression in a subset of neuroblastoma cell lines corresponds to a differentiated chromaffin cell type. Int J Cancer 2003;105:61–9. [DOI] [PubMed] [Google Scholar]
- 14.Abdallah BM, Jensen CH, Gutierrez G, et al. Regulation of human skeletal stem cell differentiation by Dlk1/Pref-1. J Bone Miner Res 2004;19:841–52. [DOI] [PubMed] [Google Scholar]
- 15.Takada S, Tevendale M, Baker J, et al. Delta-like and GTL2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol 2000;10:1135–8. [DOI] [PubMed] [Google Scholar]
- 16.Rainier S, Johnson LA, Dobry CJ, et al. Relaxation of imprinted genes in human cancer. Nature 1993;362:747–9. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa O, Eccles MR, Szeto J, et al. Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms tumour. Nature 1993;362:749–51. [DOI] [PubMed] [Google Scholar]
- 18.Charlier C, Segers K, Karim L, et al. The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat Genet 2001;27:367–9. [DOI] [PubMed] [Google Scholar]
- 19.Heathcott RW, Morison IM, Gubler MC, et al. A review of the phenotypic variation due to the Denys-Drash syndrome-associated germline WT1 mutation R362X. Hum Mutat 2002;19:462. [DOI] [PubMed] [Google Scholar]
- 20.Reeve AE, Sih SA, Raizis AM, et al. Loss of allelic heterozygosity at a second locus on chromosome 11 in sporadic Wilms’ tumor cells. Mol Cell Biol 1989;9:1799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mierau GW, Beckwith JB, Weeks DA. Ultrastructure and histogenesis of the renal tumors of childhood: an overview. Ultrastruct Pathol 1987;11:313–33. [DOI] [PubMed] [Google Scholar]
- 22.Tornehave D, Jensen CH, Teisner B, et al. FA1 immunoreactivity in endocrine tumours and during development of the human fetal pancreas; negative correlation with glucagon expression. Histochem Cell Biol 1996;106:535–42. [DOI] [PubMed] [Google Scholar]
- 23.Yin D, Xie D, De Vos S, et al. Imprinting status of DLK1 gene in brain tumors and lymphomas. Int J Oncol 2004;24:1011–15. [PubMed] [Google Scholar]
- 24.Tsibris JC, Segars J, Coppola D, et al. Insights from gene arrays on the development and growth regulation of uterine leiomyomata. Fertil Steril 2002;78:114–21. [DOI] [PMC free article] [PubMed] [Google Scholar]



