Abstract
Background: There is increasing interest in DNA methylation and in its implication in transcriptional gene silencing, a phenomenon commonly seen in human cancer.
Aims: To develop a new method that would allow quantitative DNA methylation analysis in a large range of clinical samples, independently of the processing protocol.
Methods: A methylation sensitive dot blot assay (MS-DBA) was developed, which is quantitative and combines bisulfite modification, PCR amplification using primers without CpG sites, and dot blot analysis with two probes specific for methylated and unmethylated DNA.
Results: The established method was used to study methylation of the hTERT, APC, and p16 promoter regions in microdissected, formalin fixed and paraffin wax embedded tissues.
Conclusions: MS-DBA is a sensitive, specific, and quantitative approach to analyse DNA methylation in a variety of frozen or fixed tissues. Moreover, MS-DBA is rapid, easy to perform, and permits the screening of a large panel of samples in one experiment. Thus, MS-DBA can facilitate the routine analysis of DNA methylation in all types of clinical samples.
Hypermethylation of the 5′ CpG island in gene promoter regions is a common epigenetic event in human malignancies and is associated with the transcriptional silencing of genes, notably tumour suppressor genes.1–3 The discovery of DNA methylation markers can be useful in diagnosis to identify differences between tumour and normal tissue in an individual, and also in the prognosis associated with disease progression.4 Therefore, DNA methylation based techniques are important tools in both clinical diagnostics and therapeutics. The increased interest in DNA methylation during the past few years has led to the development of many new and powerful techniques. To gain a solid understanding of the DNA methylation patterns of CpG islands in specific DNA sequences the use of bisulfite modified DNA is generally necessary.5 Bisulfite converts unmethylated cytosine to uracil, whereas methylated cytosine, present at CpG sites, remains unaltered. Several bisulfite based methods are available, each one with its own characteristics: rapidity and easiness of execution, qualitative or quantitative methylation analysis, high or low limit of methylation detection.5,6,7,8,9,10,11 However, most of these methods are not suitable for the routine analysis of clinical samples, especially formalin fixed and paraffin wax embedded tissues. Indeed, a large amount of starting DNA is generally required and a considerable number of false negative and false positive results are seen as a result of the poor quality of the extracted genomic DNA.
“Bisulfite converts unmethylated cytosine to uracil, whereas methylated cytosine, present at CpG sites, remains unaltered”
We developed a new method, the methylation sensitive dot blot assay (MS-DBA), that allows quantitative methylation analysis in a large range of clinical samples. This method is based on bisulfite treatment of DNA followed by polymerase chain reaction (PCR), using primers without CpG dinucleotides, and a dot blot analysis with two internal non-radioactive oligoprobes complementary to the amplified DNA sequences. Each probe identifies either the unmethylated or the methylated DNA sequences and reveals the proportion of cytosine or 5-methylcytosine in the DNA sample. This approach is rapid, easy to handle, and there are few false positive and false negative results. The application of MS-DBA in clinical research is wide, because this method has been applied successfully to a variety of tissues with different processing protocols encompassing frozen, archival fixed, or even microdissected formalin fixed and paraffin wax embedded tissues. Furthermore, MS-DBA permits the screening of many samples in one experiment.
MATERIALS AND METHODS
DNA extraction and sodium bisulfite conversion
Genomic DNA from stained tissue sections (microdissection) was extracted according to the standard phenol/chloroform method.12 The total amount of DNA obtained from microdissected tissues was modified in 40 μl of water with sodium bisulfite, as described previously.13
PCR amplification of hTERT, APC, and p16 gene promoters
PCR was carried out with 2 μl of bisulfite modified DNA in the presence of 5% dimethyl sulfoxide for 40 cycles with the following amplification profile: denaturation at 94°C for 30 seconds, annealing at 53°C for 45 seconds, and extension at 72°C for 75 seconds. Table 1 lists the primer sequences. To confirm correct amplification, PCR products were analysed by electrophoresis on a 2% agarose gel.
Table 1.
Primers and oligoprobes for the methylation sensitive dot blot assay
| Gene | Primer sequences (sense and antisense) | PCR product | CG sites | 3′DIG labelled oligoprobes* |
| hTERT | 5′-GGGTTATTTTATAGTTTAGGT-3′ | 224 bp | 27 | 5′-TAGTTGCGTTGTCGGGGTTA-3′ |
| 5′-AATCCCCAATCCCTC-3′ | 5′-GTAGTTGTGTTGTTGGGGTTA-3′ | |||
| APC | 5′-GGGGTTAGGGTTAGGTAGG-3′ | 196 bp | 16 | 5′-GATGCGGATTAGGGCGTTTT-3′ |
| 5′-AACTACACCAATACAACCACATA-3′ | 5′-GGATGTGGATTAGGGTGTTTT-3′ | |||
| p16 | 5′-GGGGGAGATTTAATTTGG-3′ | 194 bp | 11 | 5′-TCGGAGGGGGTTTTTTCGTT-3′ |
| 5′-CAACCCCTCCTCTTTCTT-3′ | 5′-GTTGGAGGGGGTTTTTTTGTT-3′ |
*The bold letters represent the 2 CpG sites analysed in the DNA samples.
Preparation of the positive control
SssI methylase (New England Biolabs, Beverly, Massachusetts, USA) was used to methylate 15–20 μg of normal placental or colon DNA obtained from frozen tissues. Complete methylation was confirmed by digestion with MspI (Promega, Madison, Wisconsin, USA) and HpaII (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK) restriction enzymes. SssI methylated and unmethylated DNA samples were mixed at different ratios to obtain a methylation scale (0%, 20%, 50%, 80%, and 100% of methylation). For each ratio, 2 μg of DNA was modified with sodium bisulfite and amplified by PCR for the appropriate gene as described above.
Methylation sensitive dot blot assay
MS-DBA was performed using two different oligoprobes specific to each amplified DNA sequence. Table 1 lists the probe sequences. Each probe was labelled using the DIG oligonucleotide 3′ end labelling kit (Roche, Rotkreuz, Switzerland), according to the manufacturer’s instructions. One probe contained two CG dinucleotides and was designed to recognise the methylated DNA. The other probe contained two TG dinucleotides and consequently identified the unmethylated DNA. The PCR products and the positive controls were denatured with NaOH to a final concentration of 100mM and incubated for 10 minutes at 50°C. Aliquots of 1.5 μl of the denatured PCR products were immobilised in duplicate on two NytranN membranes (Schleicher and Schuell, Dassel, Germany) and fixed with ultraviolet light for five minutes. Both membranes were prehybridised and blocked in a buffer containing 4× saline sodium citrate (SSC), 2× blocking reagent (Roche), 0.1% N-lauroylsarcosine, and 0.02% sodium dodecyl sulfate at 50°C for one hour. The solution was replaced by the hybridisation mix containing 5–10 pmol of the respective probes and the membranes were incubated for two to three hours at 50°C. The membranes were washed twice with 2× SSC at room temperature for five minutes and twice with 0.5× SSC at 53°C for 15 minutes. The membranes were blocked with maleic acid buffer containing 1× blocking reagent (Roche) for 30 minutes at room temperature and incubated with the same buffer containing 0.5 μl of anti-digoxigenin-alkaline phosphatase Fab fragments (Roche) for another 30 minutes. After washing, the reaction was developed by the addition of CDP-Star ready to use solution, according to the manufacturer’s instructions (Roche), and exposed to an X-OMAT film (Eastman Kodak Company, New York, USA). The results were read by comparing the intensity of the spots on both membranes, either visually for a semiquantitative methylation analysis or with a computer program for a quantitative analysis.
RESULTS
MS-DBA permits quantitative methylation analysis
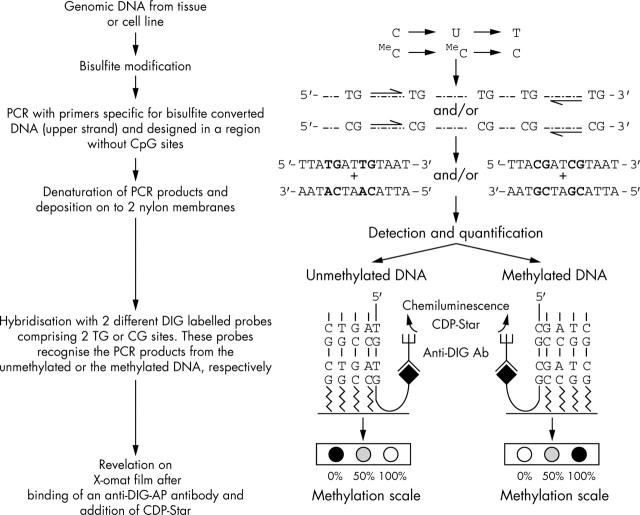
Figure 1 shows an outline of the MS-DBA method. The technique is based on bisulfite modification and PCR amplification with a dot blot readout. The combination of bisulfite modification and PCR amplification differentiates unmethylated cytosine from the methylated form. For the quantitative detection of the percentage of methylated alleles in a cell population, we used two oligoprobes labelled at their 3′ end with a DIG molecule, which were specific for the sequence of the amplified DNA. One of the probes contains two CGs (CG probe) and this probe recognises and hybridises the PCR products from the methylated DNA. The second probe contains two TGs instead of the two CGs (TG-probe) and is therefore specific for the unmethylated DNA. The number of CGs contained in the probe sequence is not an empirical condition, but two is the lower limit, because not enough hybridisation specificity will be achieved with only one CG. Different readout patterns are obtained by MS-DBA: a single dot on the membrane hybridised with the TG probe indicates no methylation (0%), a single dot on the membrane hybridised with the CG probe indicates full methylation (100%) and a dot on both membranes represents a combination of methylated and unmethylated alleles. By comparing the intensity of the methylated and unmethylated dots with an external standard, the percentage of methylated alleles can be established. Figure 2 shows the external standard obtained for the specific part of the hTERT promoter region analysed. The data show that MS-DBA yields reliable results across different ranges of DNA methylation.
Figure 1.
Outline of the methylation sensitive dot blot assay. The method is based on bisulfite modification, polymerase chain reaction amplification using primers without CpG sites and specific to the modified strand, followed by a quantitative, non-radioactive dot blot readout. The use of two DIG labelled oligoprobes specific for either the methylated or the unmethylated DNA leads to different readout patterns: no methylation, full methylation, or a combination of unmethylated and methylated alleles.
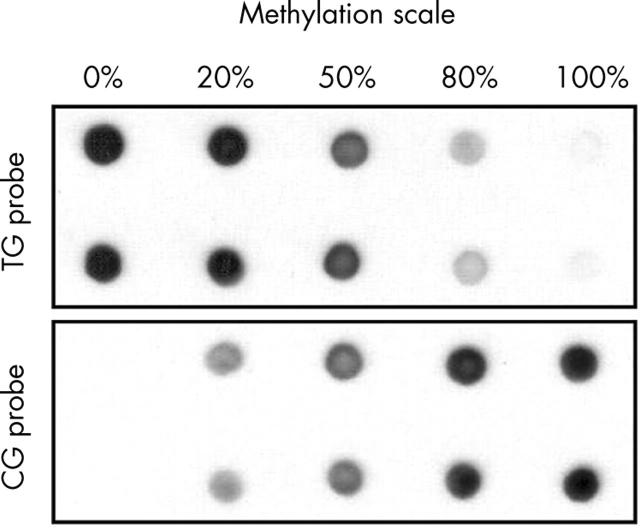
Figure 2.
Determination of quantitative methylation by methylation sensitive dot blot assay (MS-DBA) for the hTERT promoter region. DNA from normal placental tissue was methylated with SssI methylase. SssI methylated and unmethylated DNAs were mixed at different ratios as indicated above the dot blot. The data show that MS-DBA yields reliable results across a wide range of DNA methylation.
Application of MS-DBA
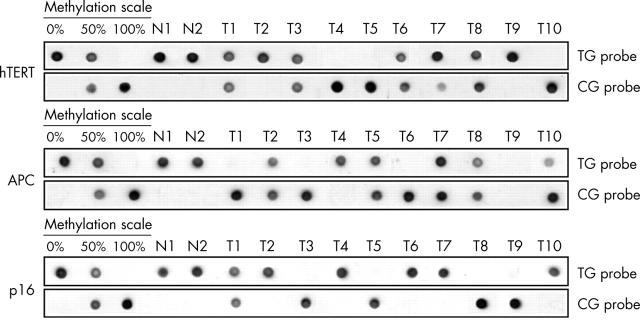
To test whether this method can be applied to the routine analysis of clinical samples, we determined the methylation status of hTERT, APC, and p16 in human oesophageal adenocarcinomas and their matched normal tissues. All the genomic DNAs were extracted after microdissection of formalin fixed and paraffin wax embedded tissue sections and found to be amplifiable by PCR after bisulfite treatment in at least 90% of cases. Reproducible results were obtained after dot blot analysis. As shown in fig 3, no methylation was found in normal oesophageal mucosal cells (N1 and N2) for the three gene promoters (hybridisation of TG probe only). The MS-DBA yielded three distinct methylation patterns in oesophageal adenocarcinoma for hTERT, APC, and p16: no methylation, full methylation (hybridisation of CG probe only), and a mixture of both (hybridisation of both probes). A mixture of both unmethylated and methylated alleles in equal amount can be explained by either methylation of the promoter in one allele with the other allele totally unmethylated, or by a heterogeneous tumorous cell population, with half of the tumour cells being fully methylated and half of the cells being unmethylated. Another possibility could arise from contamination with normal cells. However, this last point can be excluded for the cases T1, T3, T6, and T8 because full methylation was found for at least one of the three markers (fig 3), which guarantees the complete microdissection of the tumour cells. When microdissection of the lesion of interest is performed, MS-DBA gives further information about the clonality of the epigenetic alteration. For example, case T10 is fully methylated for hTERT, whereas it is only 80% methylated at APC, suggesting intratumour heterogeneity for this alteration.
Figure 3.
MS-DBA of hTERT, APC, and p16 promoter regions in human oesophageal tissues. N1 and N2, normal squamous tissues; T1–10, oesophageal adenocarcinomas. An external standard with 0%, 50%, and 100% methylation was used as control.
DISCUSSION
MS-DBA offers several advantages over existing methylation based techniques. In contrast to methylation specific PCR, the most widely used method that allows a qualitative interpretation of methylation status,6 MS-DBA provides a quantitative insight into the methylated event: it is possible to know the percentage of methylated alleles in a specific cell population within a tissue sample. The detection limit of MS-DBA was found to be less than 2% of methylated alleles in a cell population (results not shown). This allows the detection of subclones in a specific lesion.
Take home messages.
We have developed a methylation sensitive dot blot assay that is a rapid, easy to handle method for the quantitative analysis of DNA methylation in multiple samples
Because of the restricted length of the amplified product and the limitation of false positive results, this assay can be used for the routine analysis of clinical samples
DNA methylation analysis has not been extensively applied to microdissected tissue samples.13–15 The limiting amount of starting DNA necessitates appropriate PCR amplification, which increases the risk of false positive/negative results in the case of PCR over/underamplification. False positive results are also frequent with methods requiring complete bisulfite modification, such as combined bisulfite restriction analysis.10 With MS-DBA, the use of an internal oligoprobe specific to the amplified DNA fragment reduces the risk of false positive results, because no hybridisation occurs if the amplified sample is the result of a PCR artefact (fig 3; T9 for APC). It has been reported that bias during the PCR amplification can lead to an inaccurate estimation of methylation.16 Nevertheless, we generally saw linearity in the amplification of the methylated versus the unmethylated DNA if the amplified fragments did not exceed 250 bp. Furthermore, this upper limit for the PCR products is also an essential prerequisite for amplification of DNA obtained from a variety of frozen or fixed tissues.
“With the methylation sensitive dot blot assay the use of an internal oligoprobe specific to the amplified DNA fragment reduces the risk of false positive results”
MS-DBA uses the same basic method (bisulfite modification and PCR amplification) as methylation sensitive single strand conformation analysis (MS-SSCA), a semiquantitative method that can also be applied to formalin fixed, paraffin wax embedded tissues.13 Both methods use primers without CpG sites. This permits the unbiased amplification of both the methylated and unmethylated DNA. PCR products are analysed by single strand conformation analysis.13 However, reading the gel is a distinct disadvantage because PCR artefact bands can contribute to the misinterpretation of the results. Because MS-DBA PCR amplifications are performed with the same primer set used for MS-SSCA, both methods can be applied to the same sample. This could provide a double control of the results. In addition to hTERT, APC, and p16, we also tested the methylation status of the TIMP-3, RAR-β2, E-cadherin, RASSF1A, hMLH-1, and FHIT promoters in 30 cases of oesophageal adenocarcinoma by MS-SSCA and MS-DBA and found a perfect correlation between the two methods.
New DNA methylation microarray based techniques have emerged in the past few years.17,18 These methods allow methylation screening of multiple gene promoters in one sample or the mapping of individual sites of CpG methylation in genomic DNA. However, the principle of MS-DBA is not the same; it is designed to analyse one marker in a series of samples. About 40 DNA samples can be analysed in one experiment, which renders this technique very attractive for clinical studies on DNA methylation.
With the MS-DBA, we have developed a rapid, easy to handle method for the quantitative analysis of DNA methylation in multiple samples. Because of the restricted length of the amplified product and the limitation of false positive results, MS-DBA can be applied to the analysis of all tissue specimens and is therefore useful for the routine analysis of clinical samples, independently of the processing protocols.
Acknowledgments
We thank Professor FT Bosman for critical review of the manuscript. This study was supported by a grant from the Ligue Suisse Contre le Cancer (KLS-01327-02-2003).
Abbreviations
MS-DBA, methylation sensitive dot blot assay
MS-SSCA, methylation sensitive single strand conformation analysis
PCR, polymerase chain reaction
SSC, saline sodium citrate
REFERENCES
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415–28. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet 1999;21:163–7. [DOI] [PubMed] [Google Scholar]
- 3.Wajed SA, Laird PW, DeMeester TR. DNA methylation: an alternative pathway to cancer. Ann Surg 2001;234:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer 2003;3:253–66. [DOI] [PubMed] [Google Scholar]
- 5.Clark SJ, Harrison J, Paul CL, et al. High sensitivity mapping of methylated cytosines. Nucleic Acids Res 1994;22:2990–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996;93:9821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000;28:E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalgo ML, Jones PA. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nucleic Acids Res 1997;25:2529–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianco T, Hussey D, Dobrovic A. Methylation-sensitive, single-strand conformation analysis (MS-SSCA): a rapid method to screen for and analyze methylation. Hum Mutat 1999;14:289–93. [DOI] [PubMed] [Google Scholar]
- 10.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 1997;25:2532–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Maarri O, Herbiniaux U, Walter J, et al. A rapid, quantitative, non-radioactive bisulfite-SNuPE-IP RP HPLC assay for methylation analysis at specific CpG sites. Nucleic Acids Res 2002;30:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baisse B, Bian YS, Benhattar J. Microdissection by exclusion and DNA extraction for multiple PCR analyses from archival tissue sections. Biotechniques 2000;28:856–8, 860, 862. [PubMed] [Google Scholar]
- 13.Bian YS, Yan P, Osterheld MC, et al. Promoter methylation analysis on microdissected paraffin-embedded tissues using bisulfite treatment and PCR-SSCP. Biotechniques 2001;30:66–72. [DOI] [PubMed] [Google Scholar]
- 14.Lehmann U, Hasemeier B, Lilischkis R, et al. Quantitative analysis of promoter hypermethylation in laser-microdissected archival specimens. Lab Invest 2001;81:635–8. [DOI] [PubMed] [Google Scholar]
- 15.Millar DS, Warnecke PM, Melki JR, et al. Methylation sequencing from limiting DNA: embryonic, fixed, and microdissected cells. Methods 2002;27:108–13. [DOI] [PubMed] [Google Scholar]
- 16.Warnecke PM, Stirzaker C, Melki JR, et al. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res 1997;25:4422–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balog RP, de Souza YE, Tang HM, et al. Parallel assessment of CpG methylation by two-color hybridization with oligonucleotide arrays. Anal Biochem 2002;309:301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H, Maier S, Nimmrich I, et al. Oligonucleotide-based microarray for DNA methylation analysis: principles and applications. J Cell Biochem 2003;88:138–43. [DOI] [PubMed] [Google Scholar]