Abstract
Aim: To investigate the role of human papillomavirus (HPV) in the development of bladder transitional cell carcinoma (TCC).
Methods: Seventy eight paraffin wax embedded TCC samples were tested for the presence of HPV by two methods. First, immunohistochemistry was carried out using a polyclonal antibody capable of detecting the capsid protein of all known papillomaviruses. The second method was a consensus GP5+/6+ primer mediated polymerase chain reaction (PCR) technique, with the products analysed by both agarose gel electrophoresis and an enzyme immunoassay using type specific oligonucleotide probes for 10 different mucosal genotypes. To exclude false negative results because of the poor quality of DNA extracted from paraffin wax embedded samples, the series was extended to include 20 further blocks for which the corresponding snap frozen unfixed tissue was available.
Results: The two methods produced contrasting results, with 47 of the 78 samples positive for HPV antigen and none positive for HPV DNA. HPV DNA was not detected in the 20 additional paraffin wax embedded TCCs or in the 20 paired unfixed samples. In contrast, HPV DNA was amplified by PCR from all six of the paraffin wax embedded cervical carcinoma and anogenital wart control samples.
Conclusion: The disparity between the two sets of results is probably caused by false positives resulting from the non-specificity of the polyclonal antibody used for immunohistochemistry. These results suggest that HPV is unlikely to play an aetiological role in the development of bladder TCC.
Keywords: bladder cancer, transitional cell carcinoma, enzyme immunoassay-polymerase chain reaction, human papillomavirus antibody, papillomavirus
Human papillomaviruses (HPVs) are small circular DNA viruses that infect stratified squamous epithelia, namely the epidermis and mucosal epithelium. To date, more than 100 complete HPV genomes have been characterised. Although many HPV types are associated with benign lesions, certain types—such as HPV-16 and HPV-18—have an established aetiological role in tumours of the urogenital tract and anal region. HPV infection is most clearly associated with cancer of the cervix, with HPV DNA detected in over 95% of these tumours.1 HPV has also been postulated as a carcinogen in a range of other epithelial malignancies including cancers of the skin,2,3 oral cavity,4,5 oesophagus,6 and conjunctiva.7 Indeed, it has been estimated that almost 10% of the worldwide cancer burden is linked to HPV infection.8
In Western countries, more than 80% of bladder cancers are transitional cell carcinomas (TCCs), with squamous cell carcinomas the second most common morphological type identified.9 Several risk factors have been identified for urinary bladder carcinoma including environmental carcinogens, cigarette smoking, and chronic parasitic infection.10
“It has been estimated that almost 10% of the worldwide cancer burden is linked to human papillomavirus infection”
In cattle, bovine papillomavirus has been implicated in transitional cell carcinoma formation.11 Interestingly, it is the metaplastic squamous cells in these tumours that appear to contain detectable bovine papillomavirus. Previously, we have shown that squamous metaplasia is a common feature of human bladder TCC, and that its presence in muscle invasive tumours predicts a poor response to radiotherapy.11a
Several previous studies have looked for an association between HPV infection and bladder TCC development. However, the wide range of frequency detected, varying between 0% and 81%,12 has meant that the role of HPV in bladder carcinogenesis remains controversial. In our current study, we set out to examine bladder TCCs for the presence of HPV by two methods: (1) immunohistochemistry with an antibody against HPV capsid protein and (2) a polymerase chain reaction (PCR) enzyme immunoassay (EIA)13 using GP5+/6+ consensus primers, which are capable of detecting low copy number DNA from a wide spectrum of HPVs associated with mucosal epithelia.14
MATERIALS AND METHODS
Tissue samples and immunohistochemistry
In total, 98 cases of TCC were retrieved from the hospital files as paraffin wax embedded biopsies. The mean age of the patients was 73 years, with a range of 21–95. For 20 of these samples, non-fixed TCC tissue that had been snap frozen in liquid nitrogen was also available. Four paraffin wax embedded cervical carcinoma biopsies from patients with a mean age of 42.75 years, together with two paraffin wax embedded anogenital wart samples were used as positive controls for the PCR detection technique.
HPV antigen detection was performed on 5 μm thick sections of paraffin wax embedded tissue using routine immunological procedures. A polyclonal rabbit antibody against HPV capsid protein (Biogenex, San Ramon, California, USA) was used as the primary antibody at a dilution of 1/3000, with horseradish peroxidase conjugated antirabbit secondary antibody and diaminobenzidine chromogen used for visualisation of the results. HPV positive cervical carcinoma sections were used as positive controls.
DNA preparation, PCR, and sequencing
Genomic DNA was extracted from snap frozen biopsies and 5–10 μm paraffin wax embedded tissue sections using the QIAamp DNA mini kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol, with the modification that paraffin wax embedded tissue was digested with proteinase K for five days. The quality of the isolated DNA was tested by PCR amplification of the β globin gene using the primers B1 and B19.15
Mucosal HPV plasmid clones were used as positive controls. HPV types 6, 11, 16, 18, and 40 were kindly provided by E-M de Villiers (Heidelberg, Germany), HPV types 31 and 35 by A Lorincz (Gaithersburg, Maryland, USA), and HPV types 33, 34, and 39 by G Orth (Paris, France). A series of 10 fold dilutions ranging from 20 to 2 × 10−13 ng/μl was made for each plasmid DNA.
PCR amplification using GP5+/6+ biotinylated primers14 was carried out on plasmid dilutions and β globin positive clinical samples as follows. Briefly, PCR was performed in a final reaction volume of 50 μl containing either 10 μl of diluted plasmid DNA with 200 ng human placental DNA or 200 ng genomic DNA, 5 μl buffer II 10× (supplied with AmpliTaq Gold; Applied Biosystems, Foster City, California, USA), 3.5mM MgCl2, 50 μM of each deoxynucleoside triphosphate, 50 pmole of each forward and reverse primer, and 1 U of AmpliTaq Gold. Thermal cycler conditions were as follows: initial denaturation for four minutes at 94°C, followed by 40 cycles of one minute at 94°C, two minutes at 48°C, and 1.5 minutes at 72°C, with a final extension for four minutes at 72°C. Two negative controls—200 ng placental DNA and water—were included for every three genomic DNA samples. Aliquots (5 μl) from each PCR reaction were run on a 2% agarose gel, stained with 0.3 μg/ml ethidium bromide, and visualised on an ultraviolet imager.
PCR products of positive samples were gel purified using a QIAquick gel extraction kit (Qiagen) and sequenced using an ABI Prism dRhodamine terminator cycle sequencing kit (Applied Biosystems), according to the manufacturer’s protocol. Products were analysed on a Perkin Elmer 377 ABI Prism automated sequencer and BioEdit Sequence Alignment software16 was used to align the forward and reverse complement sequences. The resulting consensus sequence was compared with those of known HPV types in the GenBank database using the Blast programme on the website of the National Centre for Biotechnology Information (National Institutes of Health, Bethesda, Maryland, USA).
Enzyme immunoassay
HPV type specific oligonucleotide probes17 corresponding to the above plasmids were 3′ end labelled with digoxigenin labelled dideoxyuridine trisphosphate (DIG-ddUTP) using a DIG oligonucleotide 3′ end labelling kit (Roche Applied Sciences, Mannheim, Germany), according to the manufacturers protocol. DNA was precipitated with 0.1 volumes LiCl and six volumes of absolute ethanol to remove unincorporated DIG-ddUTP. Labelled DNA probes were resuspended in 20 μl distilled H2O and the labelling efficiency tested using a DIG nucleic acid detection kit (Roche), according to the manufacturer’s instructions.
Before their use on PCR products from clinical samples, HPV probes were first tested by EIA against GP5+/bioGP6+ PCR products from the dilution series of the relevant HPV plasmid DNA to verify sensitivity. Low risk (HPV types 6, 11, 34, and 40) and high risk (HPV types 16, 18, 31, 33, 35, and 39) HPV probes were then pooled and tested on the PCR products of LR and HR HPV plasmids to ensure the probe cocktails were group specific.
EIA was performed on GP5+/bioGP6+ PCR products from clinical samples and PCR negative controls (placental DNA and water), as described previously.18 In brief, PCR products were immobilised in a streptavidin coated microtitre plate and hybridised with probe cocktails containing 10 pmol/ml of each labelled oligonucleotide probe. Hybrids were detected by anti-DIG antibodies conjugated with alkaline phosphatase and p-nitrophenyl phosphate was used as the colorimetric substrate. Optical density was measured at 405 nm against 620 nm and a cutoff value of three times the mean optical density of the PCR negative controls was used.
RESULTS
Immunohistochemical detection of HPV capsid antigen
Immunohistochemistry was carried out on 78 paraffin wax embedded bladder TCCs. Forty seven of these samples were positive using a polyclonal antibody against HPV capsid protein (table 1; fig 1).
Table 1.
Summary of HPV detection results
| Sample | Immunohistochemistry | PCR |
| Fixed TCC | ||
| SM | 35/45 (78%) | 0/98 (0) |
| NSM | 12/33 (36%) | |
| Total | 47/78 (60%) | |
| Non-fixed TCC | ND | 0/20 (0) |
| Fixed cervical carcinoma | ND | 4/4 (100%) |
| Fixed anogenital wart | ND | 2/2 (100%) |
ND, not done; NSM, non-squamous metaplasia samples; PCR, polymerase chain reaction; SM, squamous metaplasia samples; TCC, transitional cell carcinoma.
Figure 1.
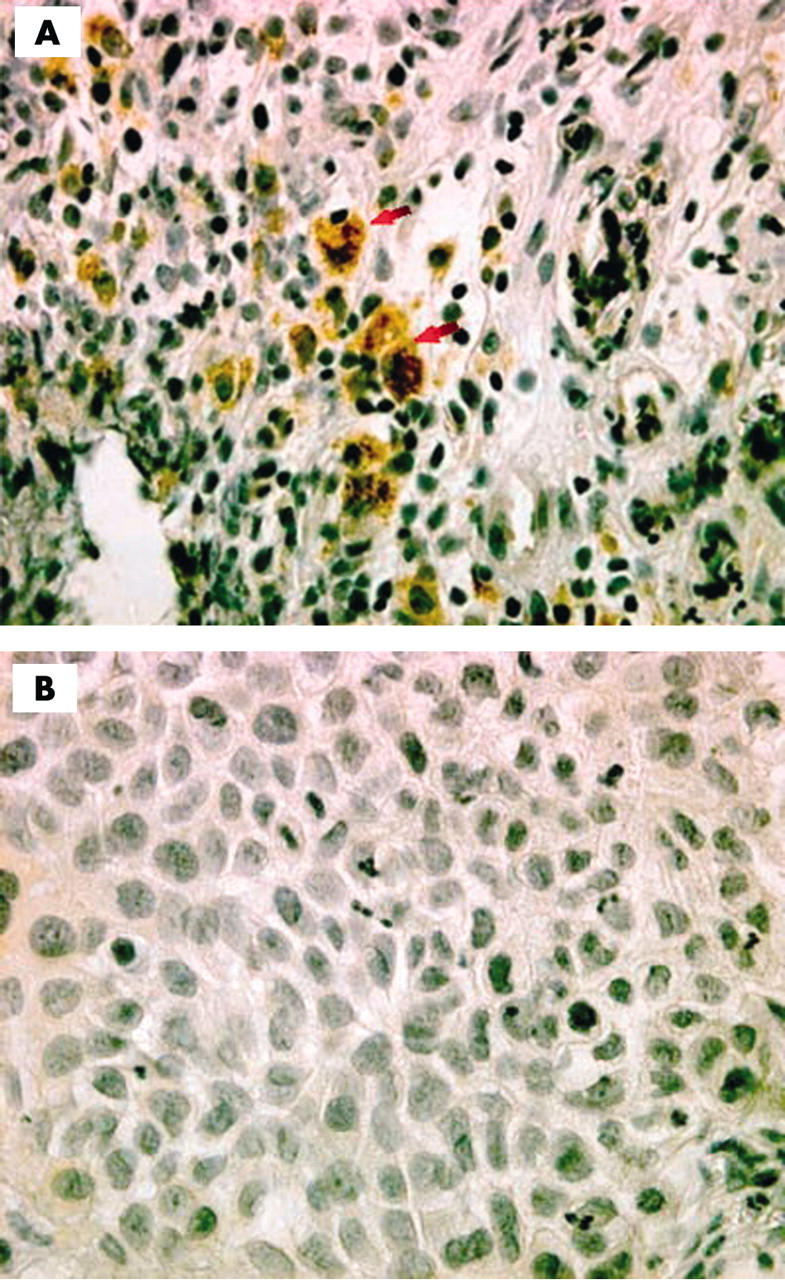
Immunohistochmical staining for human papillomavirus (HPV) capsid protein in bladder transitional cell carcinoma. Original magnification, ×40. (A) HPV positive section indicated by nuclear and cytoplasmic staining of the tumour tissue (arrowheads). (B) HPV negative section.
Detection of HPV DNA by PCR
Consensus GP5+/6+ primer mediated PCR followed by agarose gel electrophoresis was used to detect HPV DNA in 98 paraffin wax embedded bladder TCCs, 20 paired snap frozen TCC biopsies, and six positive control samples, comprising four paraffin wax embedded cervical carcinoma biopsies and two paraffin wax embedded anogenital warts. The ability of the technique to amplify HPV DNA from paraffin wax embedded tissue was demonstrated by the finding that all cervical carcinoma and anogenital wart samples were positive (table 1). The sequencing of PCR products later revealed that three of the cervical carcinoma samples were positive for HPV-16 and one was positive for HPV-18 DNA, whereas the two wart samples both contained HPV-6 DNA. In contrast, none of the paraffin wax embedded or snap frozen bladder TCCs was positive. There was no discrepancy between the results obtained using fixed and fresh frozen tissue, suggesting that the failure to detect HPV DNA in these samples was not the result of the technical difficulty of extracting good quality genomic DNA from paraffin wax embedded samples.
Screening of samples by PCR-EIA
All PCR products from clinical samples were also subjected to EIA to investigate the efficacy of PCR-EIA as a screening technique. All six of the cervical carcinoma and anogenital wart control samples in which HPV DNA was detected by PCR followed by agarose gel electrophoresis were also positive by PCR-EIA. The PCR products from the carcinoma samples were positive with the high risk but not the low risk probe cocktail, whereas the reverse was true for the wart samples, demonstrating the accuracy of EIA in distinguishing between high and low risk HPV DNA. All of the TCC samples were negative by PCR-EIA, confirming the results obtained by agarose gel electrophoresis of PCR products.
DISCUSSION
HPV has been shown to play a role in the development of anogenital cancers, particularly cervical cancer.1 In our study, we examined the possible aetiological role of HPV in bladder carcinogenesis using both immunohistochemical and PCR based detection methods. Forty seven of the 78 TCC samples screened were positive by immunohistochemistry with a polyclonal antibody against HPV L1 capsid protein. In contrast, all samples were negative for HPV DNA by PCR amplification using the consensus GP5+/6+ primer set, followed by agarose gel electrophoresis and EIA with type specific probes. The ability of the GP5+/6+ primers to amplify low copy number HPV DNA has been clearly demonstrated by de Roda Husman et al,14 who reported a detection threshold of below 100 fg HPV-16 DNA in 100 ng of human placental DNA, equivalent to less than one copy for each cellular genome. In addition, we have shown that this technique reproducibly amplified HPV DNA from all six paraffin wax embedded cervical cancer and anogenital wart control samples. Nevertheless, to exclude the possibility of false negative results as a result of the poor quality of DNA extracted from paraffin wax embedded samples, another 20 paraffin wax embedded TCCs were selected for which snap frozen tissue was also available. Negative results were also obtained after PCR amplification of DNA extracted from the fixed and frozen samples, confirming that no HPV DNA was present.
“The association of human papillomavirus with bladder transitional cell carcinoma may vary with different geographical locations”
The disparity between the above results casts doubt on the accuracy of the methods used, because it appeared unlikely that samples containing HPV DNA at levels below the PCR detection threshold would express L1 capsid protein in sufficient quantities for immunohistochemical detection. Although samples containing HPV DNA may appear as PCR false negatives if integration of the HPV genome into chromosomal DNA during carcinogenesis leads to disruption of the PCR primer binding sites, this seems unlikely to have occurred here because the GP5+/6+ primers are located within the L1 open reading frame. Indeed, we are not the first to observe a lower rate of positivity for HPV DNA compared with HPV capsid antigen in bladder TCC samples. Lopez-Beltran et al reported that 32% of 76 bladder TCCs were positive for HPV capsid antigen using a polyclonal antibody, whereas only 9% were positive for HPV DNA by PCR.19,20 These investigators suggested that the difference might have been caused by the use of a restricted range of type specific PCR primers, meaning that samples containing DNA from less common HPV types may have been overlooked. However, in view of the ability of the GP5+/6+ consensus PCR primer mediated technique to detect a range of HPV DNAs in paraffin wax embedded samples, as shown here and previously, the discrepancy between the two sets of experimental data in our present study is most probably attributable to false positives resulting from the immunohistochemical protocol, particularly because a polyclonal rather than monoclonal antibody was used.
Many earlier investigations have reported a low prevalence of HPV in TCC. For example, using a PCR methodology identical to our present study, Sur et al detected HPV DNA in only one of 64 paraffin wax embedded TCCs screened,12 and Aynaud et al found no HPV DNA in 58 bladder TCCs examined.21 Chetsanga et al detected HPV DNA in only one of 44 TCCs using a degenerate PCR technique followed by dot blot analysis with type specific probes for six HPV types commonly detected in anogenital lesions.22 Nevertheless, a few studies have reported higher incidences of HPV positivity among bladder TCCs. De Gaetani and colleagues23 detected HPV DNA in 39.5% of samples screened, whereas Chan and colleagues24 detected HPV DNA in six of 20 TCCs by dot blot analysis using type specific PCR primers and probes. Interestingly, although HPV type 6, 11, 16, 18, 31, and 33 type specific probes were used, all positive samples were found to contain HPV-18 DNA. Elsewhere, two Japanese studies25,26 found incidences of 81% and 31%, respectively. The dissimilarity in HPV prevalence reported by these investigations suggests that the association of HPV with bladder TCC may vary with different geographical locations. This is supported by the fact that most studies reporting a high incidence of HPV positive samples were performed in southern Europe or Asia,27 whereas most carried out in northern Europe and America reported an extremely low rate of HPV positivity.28 Therefore, our finding of a lack of association between HPV and bladder TCCs is not surprising.
Take home messages.
Immunohistochemistry and the polymerase chain reaction were used to investigate human papillomavirus (HPV) in transitional cell carcinoma (TCC) with very different results
Forty seven of 78 samples were positive for HPV antigen yet none was positive for HPV DNA
Because HPV DNA was not detected in unfixed TCC samples but was found in all six paraffin wax embedded cervical carcinoma and anogenital wart controls, the disparity is probably caused by the non-specificity of the polyclonal antibody used for immunohistochemistry
HPV is unlikely to play an aetiological role in the development of bladder TCC
Abbreviations
EIA, enzyme immunoassay
DIG, digoxigenin
HPV, human papillomavirus
PCR, polymerase chain reaction
TCC, transitional cell carcinoma
REFERENCES
- 1.Bosch FX, Lorincz A, Munoz N, et al. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol 2002;55:244–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGregor JM, Rustin MH. Human papillomavirus and skin cancer. Postgrad Med J 1994;70:682–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenson AB, Geyer S, Sundberg JP, et al. Human papillomavirus and skin cancer. J Investig Dermatol Symp Proc 2001;6:203–6. [DOI] [PubMed] [Google Scholar]
- 4.Barzal-Nowosielska M, Miasko A, Staroslawska E, et al. Detection of human papillomavirus in papillomas of oral cavity. Folia Histochem Cytobiol 2001;39 (suppl 2) :189–90. [PubMed] [Google Scholar]
- 5.Giovannelli L, Campisi G, Lama A, et al. Human papillomavirus DNA in oral mucosal lesions. J Infect Dis 2002;185:833–6. [DOI] [PubMed] [Google Scholar]
- 6.Syrjanen KJ. HPV infections and oesophageal cancer. J Clin Pathol 2002;55:721–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sjo NC, Heegaard S, Prause JU, et al. Human papillomavirus in conjunctival papilloma. Br J Ophthalmol 2001;85:785–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.zur Hausen H . Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta 1996;1288:F55–78. [DOI] [PubMed] [Google Scholar]
- 9.Young R . Usual variants of primary bladder lesions and secondary tumours of the bladder. In: Pathology of bladder cancer. Baltimore: Williams and Wilkins, 1996:326–37.
- 10.Malone TJ, Folberg R, Nerad JA. Lumbosacral chordoma metastatic to the eyelid. Ophthalmology 1987;94:966–70. [DOI] [PubMed] [Google Scholar]
- 11.Campo MS. Bovine papillomavirus and cancer. Vet J 1997;154:175–88. [DOI] [PubMed] [Google Scholar]
- 11a.Jenkins BJ, Martin JE, Baithun SI, et al. Prediction of response to radiotherapy in invasive cancer. Br J Urol 1990;65:345–8. [DOI] [PubMed] [Google Scholar]
- 12.Sur M, Cooper K, Allard U. Investigation of human papillomavirus in transitional cell carcinomas of the urinary bladder in South Africa. Pathology 2001;33:17–20. [PubMed] [Google Scholar]
- 13.Jacobs MV, Snijders PJ, van den Brule AJ, et al. A general primer GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J Clin Microbiol 1997;35:791–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Roda Husman AM, Walboomers JM, van den Brule AJ, et al. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol 1995;76:1057–62. [DOI] [PubMed] [Google Scholar]
- 15.Hiyama K, Kodaira M, Satoh C. Detection of deletions, insertions and single nucleotide substitutions in cloned beta-globin genes and new polymorphic nucleotide substitutions in beta-globin genes in a Japanese population using ribonuclease cleavage at mismatches in RNA:DNA duplexes. Mutat Res 1990;231:219–31. [DOI] [PubMed] [Google Scholar]
- 16.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999;41:95–8. [Google Scholar]
- 17.Jacobs MV, de Roda Husman AM, van den Brule AJ, et al. Group-specific differentiation between high- and low-risk human papillomavirus genotypes by general primer-mediated PCR and two cocktails of oligonucleotide probes. J Clin Microbiol 1995;33:901–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs MV, van den Brule AJ, Snijders PJ, et al. A non-radioactive PCR enzyme-immunoassay enables a rapid identification of HPV 16 and 18 in cervical scrapes after GP5+/6+ PCR. J Med Virol 1996;49:223–9. [DOI] [PubMed] [Google Scholar]
- 19.Lopez-Beltran A, Escudero AL, Carrasco-Aznar JC, et al. Human papillomavirus infection and transitional cell carcinoma of the bladder. Immunohistochemistry and in situ hybridization. Pathol Res Pract 1996;192:154–9. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Beltran A, Escudero AL, Vicioso L, et al. Human papillomavirus DNA as a factor determining the survival of bladder cancer patients. Br J Cancer 1996;73:124–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aynaud O, Piron D, Barrasso R, et al. Comparison of clinical, histological, and virological symptoms of HPV in HIV-1 infected men and immunocompetent subjects. Sex Transm Infect 1998;74:32–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chetsanga C, Malmstrom PU, Gyllensten U, et al. Low incidence of human papillomavirus type 16 DNA in bladder tumor detected by the polymerase chain reaction. Cancer 1992;69:1208–11. [DOI] [PubMed] [Google Scholar]
- 23.De Gaetani C, Ferrari G, Righi E, et al. Detection of human papillomavirus DNA in urinary bladder carcinoma by in situ hybridisation. J Clin Pathol 1999;52:103–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan KW, Wong KY, Srivastava G. Prevalence of six types of human papillomavirus in inverted papilloma and papillary transitional cell carcinoma of the bladder: an evaluation by polymerase chain reaction. J Clin Pathol 1997;50:1018–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anwar K, Nakakuki K, Shiraishi T, et al. Presence of ras oncogene mutations and human papillomavirus DNA in human prostate carcinomas. Cancer Res 1992;52:5991–6. [PubMed] [Google Scholar]
- 26.Furihata M, Ohtsuki Y, Ogoshi S, et al. Prognostic significance of human papillomavirus genomes (type-16, -18) and aberrant expression of p53 protein in human esophageal cancer. Int J Cancer 1993;54:226–30. [DOI] [PubMed] [Google Scholar]
- 27.Yu Z, Xia T, Xue Z. [The detection of high risk human papillomaviruses in papillary transitional cell carcinoma of urinary bladder.] Zhonghua Wai Ke Za Zhi 1999;37:369–71. [PubMed] [Google Scholar]
- 28.Simoneau M, LaRue H, Fradet Y. Low frequency of human papillomavirus infection in initial papillary bladder tumors. Urol Res 1999;27:180–4. [DOI] [PubMed] [Google Scholar]


