Abstract
Aims: To develop a quantitative reverse transcriptase polymerase chain reaction (Q-RT-PCR) for severe acute respiratory syndrome coronavirus (SARS-CoV) detection and explore the potential of using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA as an internal control to exclude false negative results.
Methods: SARS-CoV and GAPDH mRNA were both measured in 26 specimens from 16 patients with SARS, 40 follow up specimens from the same batch of patients, and appropriate control subjects. The relation between SARS positivity and GAPDH mRNA concentration was investigated using the χ2 test. Increasing the sensitivity for SARS-CoV and GAPDH mRNA detection was investigated in follow up specimens in which SARS-CoV and GAPDH mRNA were not detected initially.
Results: Varying amounts of SARS-CoV were found in the 26 SARS-CoV positive specimens and SARS-CoV was not detected in the 40 follow up specimens and controls. In addition, concentrations of GAPDH mRNA were significantly different between the patients with SARS, follow up specimens, and healthy controls (Kruskal-Wallis test, p<0.05). Moreover, GAPDH mRNA concentrations were highly correlated with SARS-CoV positivity (χ2 = 5.43; p<0.05). Finally, SARS-CoV and GAPDH mRNA were both detected in three follow up urine specimens that were initially negative when the amount of cDNA used was increased from 5 μl to 10 and 15 μl.
Conclusions: This Q-RT-PCR assay can be used to detect SARS-CoV. Moreover, GAPDH mRNA may be useful to rule out false negative results in SARS-CoV detection, and the current extraction method for urine may not be sensitive enough to detect low titres of SARS-CoV.
Severe acute respiratory syndrome (SARS) is a new infectious disease with high transmissibility in healthcare settings and an international spread through travel.1,2 In total, 8437 probable SARS cases were reported to the World Health Organisation (WHO) from 29 countries between November 1 2002 and July 11 2003, with 813 deaths (mortality rate, 9.6%).1 The causative agent is a novel coronavirus (CoV), which had been isolated from Himalayan palm civets in Guangdong, China.3–5
“Molecular techniques have drawbacks arising from the lack of standardisation in sampling techniques and extraction protocols for various types of body fluids, which may lead to false negative results in some specimens”
Molecular tests have been developed for the detection of SARS-CoV during the acute phase of the disease in various respiratory samples, such as nasopharyngeal swabs, nasal swabs, nasopharyngeal aspirates, throat swabs, sputum, bronchial alveolar lavage specimens, stool, urine, and serum.6–8 The most commonly used technique for the molecular detection of SARS-CoV is the reverse transcriptase polymerase chain reaction (RT-PCR), but the assay cannot provide data for quantitative analysis and is potentially less sensitive than quantitative RT-PCR (Q-RT-PCR). In addition, there are drawbacks arising from the lack of standardisation in sampling techniques and extraction protocols for various types of body fluids, which may lead to false negative results in some specimens. In this report, we describe a Q-RT-PCR assay, using the recently released genomic sequences9 for SARS-CoV detection, and explore the possibility of using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, which is commonly used to standardise non-homogeneous body fluid specimens,10–12 as a surrogate marker to exclude false negative results. The information obtained would be useful to help us understand the relation between SARS-CoV and GAPDH mRNA in various kinds of clinical specimens.
MATERIALS AND METHODS
Ethics
Our study was approved by the local institutional research ethics committee. Permission for obtaining data from hospital records was granted by the department of pathology, Queen Elizabeth Hospital, Hong Kong Special Administrative Region, China.
Specimens
Twenty six clinical specimens, which had tested positive for SARS-CoV by the conventional RT-PCR test, were retrieved retrospectively. All specimens except stool were received in 2 ml of viral culture medium. The specimens were from 16 patients (table 1) admitted to Queen Elizabeth Hospital, Hong Kong, China, between April and June 2003, who fulfilled the WHO case definition of SARS. The specimens comprised nine nasopharyngeal swabs, two nasal swabs, five rectal swabs, one throat swab, one nasopharyngeal aspirate, three tracheal aspirates, and five stool samples. The second group comprised 40 RT-PCR negative specimens collected from the above 16 patients with SARS, during their follow up consultation, after discharge from hospital for two to 12 weeks. There were 13 nasopharyngeal swabs, 12 stool samples, eight urine samples, five throat swabs, one rectal swab, and one tracheal aspirate specimen. A control group, comprising throat swabs from 40 patients with unrelated diseases composed of common cold (12), chronic bronchitis (five), asthma (three), diabetes (eight), gastric ulcer (two), anaemia (three), and rheumatoid arthritis (seven) was used to evaluate the specificity of the assay. For GAPDH mRNA detection, throat swabs from another 40 apparently healthy subjects were used as controls.
Table 1.
Clinical data and specimen types of SARS-CoV positive patients
| Patient | Sex | Age (years) | Specimen type | SARS-CoV (copies/ml) | Days of onset |
| 1 | F | 31 | Nasopharyngeal swab | 24 700 | 30 |
| 2 | F | 26 | Nasopharyngeal swab | 676 000 | 6 |
| Rectal swab | 1780 000 000 | 7 | |||
| Tracheal aspirate | 60 100 | 21 | |||
| Nasopharyngeal aspirate | 57 800 | 21 | |||
| 3 | F | 42 | Nasopharyngeal swab | 50 000 000 | 17 |
| 4 | M | 38 | Rectal swab | 51 500 000 | 19 |
| Tracheal aspirate | 810 000 | 18 | |||
| 5 | M | 79 | Throat swab | 653 000 | 21 |
| Stool | 77 400 | 21 | |||
| Stool | 1200 000 000 | 23 | |||
| 6 | M | 23 | Nasal swab | 205 000 | 16 |
| 7 | M | 30 | Nasopharyngeal swab | 5150 000 | 16 |
| 8 | M | 46 | Nasopharyngeal swab | 69 700 | 56 |
| 9 | F | 36 | Nasal swab | 149 000 | 28 |
| 10 | F | 39 | Stool | 3390 000 | 5 |
| Nasopharyngeal swab | 52 500 000 | 11 | |||
| Nasopharyngeal swab | 69 700 000 | 17 | |||
| Nasopharyngeal swab | 5300 000 | 23 | |||
| 11 | F | 55 | Rectal swab | 3060 000 000 | 14 |
| 12 | F | 75 | Tracheal aspirate | 93 200 000 | 21 |
| 13 | F | 26 | Rectal swab | 6430 000 | 23 |
| Nasopharyngeal swab | 108 000 | 34 | |||
| 14 | F | 55 | Rectal swab | 31 500 000 | 8 |
| 15 | F | 31 | Stool | 561 000 | 11 |
| 16 | F | 29 | Stool | 19 000 000 | 11 |
SARS-CoV, severe acute respiratory syndrome coronavirus.
RNA extraction and reverse transcription
All specimens except stool were mixed thoroughly in viral culture medium before 140 μl aliquots were used for RNA extraction according to the mini spin protocol of the QIAamp virus RNA mini kit (Qiagen, Hilden, Germany), which can bind all RNA molecules greater than 200 nucleotides in length. The extracted RNA was eluted in 60 μl RNase free water, treated with deoxyribonuclease I (Life Technologies, Carlsbad, California, USA) to digest genomic DNA, and stored at −80°C before use. For stool samples, 5 ml phosphate buffered saline was added and RNA was extracted from 140 μl of the resulting suspension as above. Reverse transcription was performed using the TaqMan Gold RT-PCR kit (Applied Biosystems, Foster City, California, USA), which has a high efficiency—with conversion of one copy of RNA to one copy cDNA—according to the recommended protocol. cDNA was generated in a 20 μl reaction mix, and stored at −20°C until use. Each batch of reactions included positive controls for SARS-CoV or GAPDH mRNA and negative controls without reverse transcriptase.
Q-RT-PCR assay
Specific primers and fluorescent probes for SARS-CoV and GAPDH mRNA detection (as described previously by us13) were purchased from Applied Biosystems (table 2). The SARS-CoV primers and probe perfectly matched all available genomic sequences on GenBank. The primers and probe targeted the open reading frame 1ab region (accession number, AY313906: nucleotides 15336–15357 and 15387–15414 for forward and reverse primers, respectively). Moreover, the specificity of the primers and probe was checked on the BLAST program (NCBI) and showed no reactivity other than SARS-CoV. GAPDH mRNA detection was highly specific because of its intron spanning nature. Q-RT-PCR was set up in a reaction volume of 50 μl using the Core Reagents kit (Applied Biosystems) and a standard protocol with 40 cycles on the ABI Prism 7000 Sequence Detector (Applied Biosystems). For the SARS-CoV positive patients and the follow up specimens, 5 μl aliquots of cDNA were used for both SARS-CoV and GAPDH mRNA detection. To increase the sensitivity of the assay in follow up specimens that were negative when first tested, the amount of cDNA used was increased 10 μl and 15 μl for both SARS-CoV and GAPDH. To confirm the GAPDH mRNA results, we also measured β actin mRNA using a predesigned Taqman probe and intron spanning primers (Applied Biosystems) in the same batch of specimens.
Table 2.
Sequences of primers, probes, and calibration oligonucleotide
| Primer/probe | Sequence |
| SARS-CoV | |
| Primers | |
| Forward | 5′-AGCTAACGAGTGTGCGCAAGTA-3′ |
| Reverse | 5′-TGATGTTCCACCTGGTTTAACATATAGT-3′ |
| Dual labelled fluorescent probe | 5′-(FAM*)AGCCGCCACACATGACCATCTCACTTA(TAMRA†)-3′ |
| Oligonucleotide for calibration | 5′-AGGTTAGCTAACGAGTGTGCGCAAGTATTAAGTG AGATGGTCATGTGTGGCGGCTCACTATATGTTAAACCAGGT GGAACATCATCCGG-3′ |
| GAPDH mRNA | |
| Primers | |
| Forward | 5′-GAAGGTGAAGGTCGGAGT-3′ |
| Reverse | 5′-GAAGATGGTGATGGGATTTC-3′ |
| Dual labelled fluorescent probe | 5′-(FAM*)CAAGCTTCCCGTTCTCAGCC(TAMRA†)-3′ |
*6-Carboxyfluorescein; †6-carboxytetramethylrhodamine.
The calibration curve for the measurement of SARS-CoV was prepared by amplifying serial dilutions of a synthetic oligonucleotide (table 2), with concentration ranging from 25 to 2.5 × 106 copies/μl. Calibration curves for GAPDH and β actin mRNA were prepared by serial dilutions of cDNA reverse transcribed from human control RNA (Applied Biosystems), with concentrations ranging from 41 to 2.6 × 105 copies/μl, based on the manufacturer’s information that 1 pg of control RNA contained approximately 100 copies of GAPDH and β actin transcripts. Each amplification batch included positive controls for SARS-CoV, GAPDH, and β actin, calibration oligonucleotides, and a negative control without cDNA. Duplicate tests were performed and the average was calculated for each sample.
Statistical analysis
The Kruskal-Wallis test was used to determine whether there was a significant difference in the concentration of GAPDH and β actin mRNA between the different groups of specimens. The χ2 test was used to investigate the relation between GAPDH mRNA concentrations and SARS positivity. GraphPad Prism software version 4.0 (GraphPad Software Inc, 1999–2003; San Diego, California, USA) was used for all statistical analysis and p < 0.05 was considered significant.
RESULTS
Development of Q-RT-PCR assay
Because the ABI 7000 sequence detector uses a tungsten detection system rather than a laser capture system, it has a lower detection limit of approximately 100 copies. Thus, only copy numbers over 100 are interpreted as being positive. The calibration curve was not extrapolated, so that all positive results were within the range detected by calibration standards in each Q-RT-PCR assay.
Treatment of raw data and quality control
In each extraction, a 140 μl sample was loaded on to the column and RNA was finally eluted in 60 μl. Subsequently, 7.6 μl RNA in a 20 μl reaction mix was used in each reverse transcription reaction. Finally, 5 μl cDNA was used in each Q-RT-PCR assay. Therefore, the dilution factor was 4.0 × 7.9 × 7.1 = 224.4. All positive controls had the predicted quantities of SARS-CoV and GAPDH mRNA. In all assay batches the negative controls showed no signals.
SARS-CoV detection in confirmed SARS, SARS follow up specimens, and patients with unrelated diseases
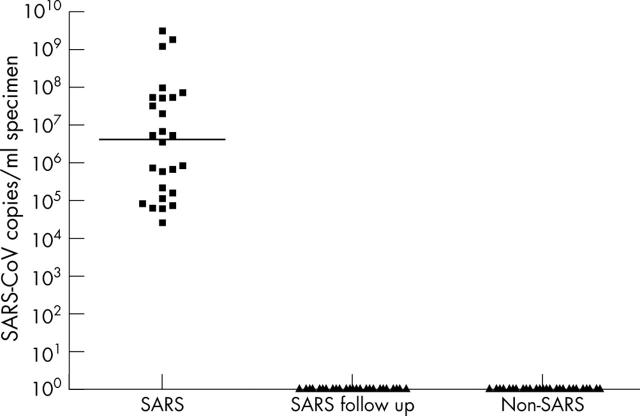
Varying amounts of SARS-CoV were found in the 26 RT-PCR positive samples from the 16 patients with SARS (table 1), with titres ranging from 2.47 × 104 to 3.06 × 109 copies/ml. SARS-CoV was not detected in the 40 SARS follow up specimens or the specimens from patients with unrelated diseases (fig 1).
Figure 1.
Severe acute respiratory syndrome coronavirus (SARS-CoV) copies/ml in SARS-CoV positive patients, follow up specimens, and patients with unrelated diseases. The median in each group of subjects is indicated by the black horizontal line.
GAPDH mRNA detection in confirmed SARS, SARS follow up specimens, and healthy controls
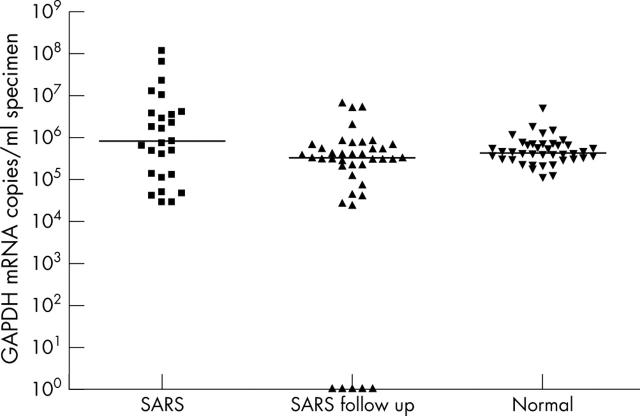
Varying concentrations of GAPDH mRNA were found in all specimens, except for five follow up cases that had no detectable GAPDH mRNA (fig 2). The median GAPDH mRNA concentrations were 8.2 × 105 (range, 3.0 × 104 to 1.2 × 108), 3.4 × 105 (range, 0 to 7.1 × 106), and 4.5 × 105 (range, 1.2 × 105 to 5.2 × 106) in SARS positive patients, SARS follow up specimens, and apparently healthy subjects, respectively. Statistical analysis indicated that there was a significant difference in GAPDH mRNA concentrations between these three groups (Kruskal-Wallis test, p < 0.05).
Figure 2.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA copies/ml in specimens from severe acute respiratory syndrome (SARS) coronavirus positive patients, follow up specimens, and apparently normal subjects. The median in each group of subjects is indicated by the black horizontal line.
Relation between GAPDH mRNA concentration and SARS-CoV positivity
Samples from SARS positive patients and follow up specimens were divided into two groups: those with a high GAPDH mRNA concentration (> 1 × 106 copies/ml) and those with a low GAPDH mRNA concentration (< 1 × 105 copies/ml). This stratification was entirely arbitrary: only 24% of samples had a GAPDH mRNA concentration > 1 × 106 copies/ml and only 23% had < 1 × 105 copies/ml, so that 53% of the samples were in between these two values. In the high GAPDH mRNA concentration group, there were 12 SARS positive cases, whereas there were only four SARS negative cases. Similarly, in the low GAPDH mRNA concentration group, there were five SARS positive cases and 10 SARS negative cases. The χ2 test revealed a highly significant association between GAPDH mRNA concentration and SARS-CoV positivity (χ2 = 5.43; p < 0.05).
Increased sensitivity for SARS-CoV and GAPDH mRNA in follow up specimens using higher amounts of cDNA
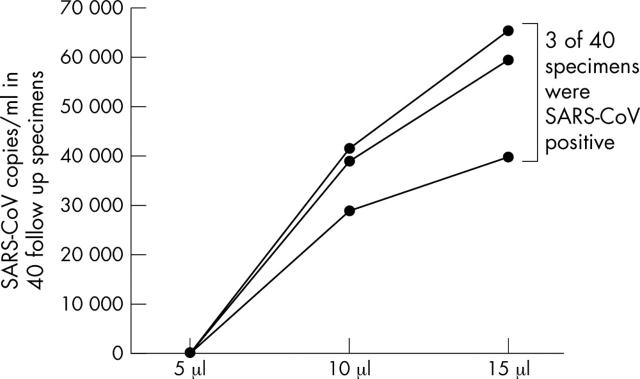
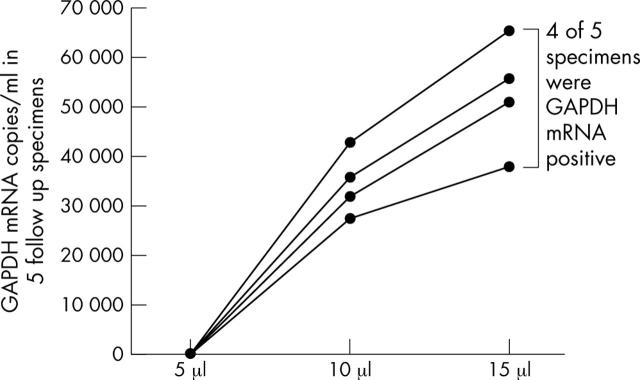
SARS-CoV could be identified in three of 40 follow up specimens when 10 μl and 15 μl cDNA aliquots were used (fig 3), whereas the other 37 specimens remained negative even when these higher amounts of cDNA were used. In addition, GAPDH mRNA was found in four of five initially negative follow up specimens when 10 μl and 15 μl aliquots of cDNA were used (fig 4). Surprisingly, the three specimens that changed to being SARS-CoV positive when higher amounts of cDNA were used (all urine specimens) also became GAPDH mRNA positive.
Figure 3.
Severe acute respiratory syndrome coronavirus (SARS-CoV) copies/ml in 40 follow up specimens when 5 μl, 10 μl, and 15 μl aliquots of cDNA were used in the quantitative reverse transcriptase polymerase chain reaction assay.
Figure 4.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA copies/ml in five follow up specimens when 5 μl, 10 μl, and 15 μl aliquots of cDNA were used in the quantitative reverse transcriptase polymerase chain reaction assay.
Confirmation of GAPDH mRNA results by detecting β actin mRNA
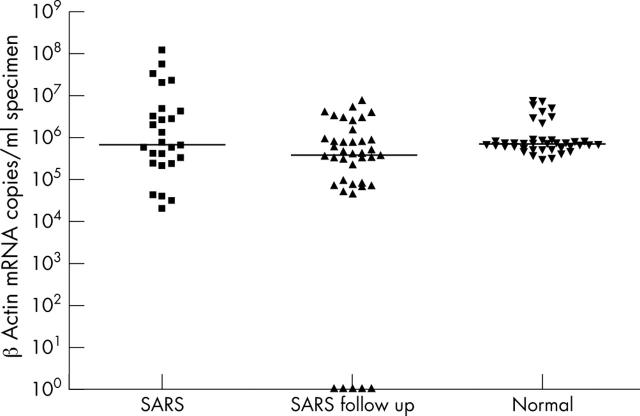
A similar pattern, with significant differences between patients with confirmed SARS, SARS follow up specimens, and apparently healthy controls, was seen for β actin mRNA (fig 5; Kruskal-Wallis test, p < 0.05). Moreover, β actin mRNA was detected in four of five follow up specimens when 10 and 15 μl aliquots of cDNA were used, although it was not detected when 5 μl was used initially. As expected, three of these four specimens were the same urine samples that also became positive in the SARS-CoV and GAPDH assays when higher amounts of cDNA were used.
Figure 5.
β Actin mRNA copies/ml in severe acute respiratory syndrome (SARS) coronavirus positive patients, follow up specimens, and apparently normal subjects. The median in each group of subjects is indicated by the black horizontal line.
DISCUSSION
The successful demonstration of SARS-CoV in all patients with confirmed SARS and its absence in controls with unrelated diseases clearly attests to the suitability of the Taqman probe and primers used in this assay and to the optimisation of the protocol. This method provides a sensitive Q-RT-PCR assay, which is much less expensive than other commercially available kits, for the detection of SARS-CoV in clinical laboratories.
The measurement of GAPDH mRNA concentrations in our study provided vital information that may be beneficial to SARS-CoV detection in the future.
First, there was a significant difference in GAPDH mRNA concentrations between the specimens from patients with SARS during their illness, those from patients during follow up, and those from apparently healthy controls. We think that there are two possible reasons for this variation. One is that cellular activity may be increased in patients with SARS during their illness compared with during recovery and in health. This speculation is based on recent reports that GAPDH is involved in many diverse cellular functions unrelated to glycolysis, including nuclear RNA export, DNA replication, DNA repair, exocytotic membrane fusion, apoptosis, neurodegenerative disease, prostate cancer, and viral pathogenesis.14,15 Another reason is that the absence of detectable GAPDH mRNA in five follow up specimens (two nasopharyngeal swabs and three urine samples) (fig 2) would lower the overall GAPDH mRNA concentration in the follow up specimens. These five samples were useful for investigating the potential use of GAPDH mRNA as an internal control to exclude false negative results in SARS-CoV detection.
Second, a much broader range in GAPDH mRNA concentrations in patients with active SARS and in patients during follow up compared with apparently normal subjects may reflect the importance of the standardisation of sampling techniques because only one clinician was responsible for taking the specimens from the apparently healthy subjects, whereas more than eight were involved in taking specimens from each of the two groups of patients tested.
Take home messages.
The quantitative reverse transcriptase polymerase chain reaction assay described here can be used to detect severe acute respiratory syndrome coronavirus (SARS-CoV)
Glyceraldehyde-3-phosphate dehydrogenase mRNA may be a useful internal marker to rule out false negative results in SARS-CoV detection
The current extraction method for urine may not be sensitive enough to detect low titres of SARS-CoV, and further investigations are needed
“The ultimate aim of using GAPDH mRNA should be to establish a baseline or a reference range below which a negative severe acute respiratory syndrome coronavirus result may not be reliable”
Third, the χ2 test showed that, in general, SARS positive specimens had higher GAPDH mRNA concentrations whereas SARS negative specimens had lower GAPDH mRNA concentrations. This significant association indicates that false negative SARS-CoV results may be found in those specimens with low GAPDH mRNA concentrations, as demonstrated by the lack of detectable GAPDH mRNA and SARS-CoV in five follow up specimens when only 5 μl of cDNA was used.
Although there are conflicting reports on the use of GAPDH mRNA for normalising mRNA values in Q-RT-PCR assays,10–12,14,16 the successful detection of SARS-CoV, GAPDH, and even β actin mRNA in three follow up urine specimens with increased amounts of cDNA reaffirms the importance of using GAPDH mRNA as an indicator of false negative results. We appreciate the fact that the GAPDH mRNA concentration can vary significantly among different individuals,17 and during pregnancy,18 oxidative stress,19 hypoxia,20 and cancer,21 so that GAPDH mRNA may not be suitable to normalise SARS-CoV concentrations. Therefore, the ultimate aim of using GAPDH mRNA should be to establish a baseline or a reference range below which a negative SARS-CoV result may not be reliable. The detection of GAPDH and β actin mRNA was not the result of expression from pseudogenes because DNase had been added to each sample to digest any genomic DNA present, and also because the sequence of five amplified products from each of these genes had been identified to confirm the specificity of the assay (data not shown).
The successful detection of SARS-CoV and GAPDH mRNA in three of eight urine specimens after increasing the amount of cDNA used in the Q-RT-PCR assay shows that the current urine extraction protocol may not be sensitive enough because urine specimens have a very broad range of concentration, which depends on the patients’ water intake and renal function. This area demands further investigation. Thus, our study provides another choice of Taqman probe and primers to be used in Q-RT-PCR and is the first to look at the correlation between GAPDH mRNA and SARS-CoV in clinical specimens. The development of a multiplex assay for both SARS-CoV and GAPDH mRNA may be essential to rule out false negative results in the future. Because asymptomatic and subclinical SARS-CoV infection may exist in the community, as shown in the recently confirmed case by the WHO in Guangdong, China, a more sensitive detection protocol and appropriate samples will enable us to detect this virus at the earliest possible time, so that even the slightest chance of spreading can be prevented in the future.
Acknowledgments
We thank all the health care workers who were involved in managing the patients with SARS and the specimens during the SARS outbreak in Hong Kong. The support of the Pathology Department, Queen Elizabeth Hospital is gratefully appreciated.
Abbreviations
cDNA, complementary deoxyribonucleic acid
CoV, coronavirus
GAPDH, glyceraldehyde-3-phosphate dehydrogenase
Q-RT-PCR, quantitative reverse transcriptase polymerase chain reaction
RT-PCR, reverse transcriptase polymerase chain reaction
SARS, severe acute respiratory syndrome
WHO, World Health Organisation
REFERENCES
- 1.World Health Organisation. Cumulative number of reported cases of severe acute respiratory syndrome (SARS) (available at http://www.who.int/csr/sars/country/2003_07_11.en/).
- 2.Bell D, Jenkins P, Hall J. World Health Organisation global conference on severe acute respiratory syndrome. Emerg Infect Dis 2003;9:1191–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. SARS coronavirus sequencing (available at: http://www.cdc.gov/ncidod/sars/sequence.htm).
- 4.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003;361:1319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science 2003;302:276–8. [DOI] [PubMed] [Google Scholar]
- 6.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967–76. [DOI] [PubMed] [Google Scholar]
- 7.Poon LL, Chan KH, Wong OK, et al. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J Clin Virol 2003;28:233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng EK, Hui DS, Chan KC, et al. Quantitative analysis and prognostic implication of SARS coronavirus RNA in the plasma and serum of patients with severe acute respiratory syndrome. Clin Chem 2003;49:1976–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsui SK, Chim SS, Lo YM, et al. Coronavirus genomic-sequence variations and the epidemiology of the severe acute respiratory syndrome. N Engl J Med 2003;349:187–8. [DOI] [PubMed] [Google Scholar]
- 10.Gueudin M, Vabret A, Petitjean J, et al. Quantitation of respiratory syncytial virus RNA in nasal aspirates of children by real-time RT-PCR assay. J Virol Methods 2003;109:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy CL, Polak JM. Differentiating embryonic stem cells: GAPDH, but neither HPRT nor beta-tubulin is suitable as an internal standard for measuring RNA levels. Tissue Eng 2002;8:551–9. [DOI] [PubMed] [Google Scholar]
- 12.Juusola J, Ballantyne J. Messenger RNA profiling: a prototype method to supplant conventional methods for body fluid identification. Forensic Sci Int 2003;135:85–96. [DOI] [PubMed] [Google Scholar]
- 13.Ng EK, Tsui NB, Lam NY, et al. Presence of filterable and nonfilterable mRNA in the plasma of cancer patients and healthy individuals. Clin Chem 2002;48:1212–17. [PubMed] [Google Scholar]
- 14.Tatton WG, Chalmers-Redman RM, Elstner M, et al. Glyceraldehyde-3-phosphate dehydrogenase in neurodegeneration and apoptosis signaling. J Neural Transm Suppl 2000;60:77–100. [DOI] [PubMed] [Google Scholar]
- 15.Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta 1999;1432:159–84. [DOI] [PubMed] [Google Scholar]
- 16.Glare EM, Divjak M, Bailey MJ, et al. Beta-actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax 2002;57:765–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bustin SA, Gyselman VG, Williams NS, et al. Detection of cytokeratins 19/20 and guanylyl cyclase C in peripheral blood of colorectal cancer patients. Br J Cancer 1999;79:1813–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cale JM, Millican DS, Itoh H, et al. Pregnancy induces an increase in the expression of glyceraldehyde-3-phosphate dehydrogenase in uterine artery endothelial cells. J Soc Gynecol Investig 1997;4:284–92. [PubMed] [Google Scholar]
- 19.Ito Y, Pagano PJ, Tornheim K, et al. Oxidative stress increases glyceraldehyde-3-phosphate dehydrogenase mRNA levels in isolated rabbit aorta. Am J Physiol 1996;270:H81–7. [DOI] [PubMed] [Google Scholar]
- 20.Zhong H, Simons JW. Direct comparison of GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem Biophys Res Commun 1999;259:523–6. [DOI] [PubMed] [Google Scholar]
- 21.Chang TJ, Juan CC, Yin PH, et al. Up-regulation of beta-actin, cyclophilin and GAPDH in N1S1 rat hepatoma. Oncol Rep 1998;5:469–71. [DOI] [PubMed] [Google Scholar]