Abstract
Aims: To investigate the incidence of genetic aberrations in the DNA repair genes in a cohort of oesophageal cancers.
Methods: One hundred oesophagectomy samples of squamous cell carcinoma were studied. Normal and tumour DNA were isolated using a standard phenol/chloroform extraction procedure. Six recommended microsatellite loci with high informativity were analysed. The following markers were used: D2S123 (2p), D3S659 (3p), D3S1255 (3p), Bat 25 (4q), Bat 26 (2p), and Bat 40 (1p). The results were analysed using software attached to an automated DNA sequencer. The molecular data were then correlated with clinicopathological parameters.
Results: The incidence of microsatellite instability and loss of heterozygosity was very low. There was no significant correlation between the clinicopathological and molecular data. However, D2S123 genetic abnormalities were seen more frequently in both moderately and well differentiated tumours than in poorly differentiated tumours (p = 0.033). Follow up data were available for only 67 of the 100 patients. Fifty patients were alive and 17 patients had died.
Conclusion: Low frequencies of genetic aberrations in these mismatch repair loci are found in squamous carcinomas of the oesophagus from a high incidence area in South Africa.
Keywords: BAT 25, DNA repair genes, fluorescence based technology, oesophageal cancer, microsatellite polymerase chain reaction
Oesophageal cancer is the most common malignancy in black South African men, accounting for 14.3% of all cancers in South Africa.1 Over and above the various environmental factors that impact on the development of cancer in general, great advances have been made during the past decade in our understanding of the molecular mechanisms that occur in the transition from normal mucosa, through dysplasia, and finally to carcinoma.2–4 Several studies have revealed that an accumulation of genetic alterations gives rise to cancer. Furthermore, microsatellite instability (MSI) and loss of heterozygosity (LOH) at highly informative genetic loci at which tumour suppressor genes and oncogenes are located are thought to play a role in carcinogenesis.5,6,7,8,9,10 Genes investigated in recent studies include p53, Rb, DCC, APC, and MCC.11 We recently showed that the occurrence of MSI in South African squamous carcinomas of the oesophagus was similar to other high incidence areas.7
“The mismatch repair genes play an important role in ensuring that errors occurring during DNA synthesis are corrected”
However, very little is known about the status of the mismatch repair genes and their impact on oesophageal cancer. The mismatch repair genes play an important role in ensuring that errors occurring during DNA synthesis are corrected. These genes were initially identified in bacterial systems and mutations in DNA mismatch repair genes have been associated with hereditary non-polyposis colorectal cancer.12–14 Five human DNA mismatch repair genes have been identified, which when mutated cause susceptibility to both sporadic and hereditary non-polyposis colorectal cancer. Mutational inactivation of both copies of a mismatch repair gene results in a profound repair defect and progressive accumulation of mutations throughout the genome. Our study focuses on evaluating the incidence of genetic aberrations in the mismatch repair genes and its clinical impact on the development of oesophageal cancer in a high incidence area of South Africa.
MATERIALS AND METHODS
One hundred oesophagectomy specimens were obtained from the department of pathology, Nelson R Mandela School of Medicine, Durban, South Africa. The specimens were processed for routine histological examination after fixing in 10% buffered formal saline. Sections were stained with haematoxylin and eosin and all cases were reviewed by one of the authors (RC). Cases were graded as well, moderately, and poorly differentiated. Staging was performed using the TNM/UICC system, and clinical follow up was obtained from the department of cardiothoracic surgery. Normal and tumour sections were selected for DNA extraction after histological evaluation. Only sections containing more than 80% tumour tissue were used for extraction of tumour DNA. These areas were microdissected from the surrounding normal tissue.
DNA extraction
DNA was extracted from formalin fixed, paraffin wax embedded tissue blocks. Three sections (6 μm thick) were cut from the paired normal and tumour blocks. The procedure used was essentially as described by Naidoo et al.15 The quality of the isolated DNA was verified in a standard polymerase chain reaction (PCR) using primers targeting the ubiquitous insulin gene.16
Microsatellite PCR
CY5 labelled primers
The microsatellite primers were purchased from Roche Diagnostics, Penzberg, Germany. Table 1 lists the primer sequences for the mismatch repair markers used in our study. These markers were chosen because of the recommendation of the American Association for Cancer Research.17 They are also deemed to be highly informative and significant when investigating mismatch repair abnormalities.
Table 1.
Sequence data for the microsatellite markers
| Marker | Primer sequence | Size range (bp) | Annealing temperature (°C) |
| D2S123 | (F) 5′-AAACAGGATGCCTGCCTTTA-3′ | 197–227 | 55 |
| (R) 5′-GAACTTTCCACCTATGGGAC-3′ | |||
| D3S659 | (F) 5′-ATTCCAGGGACAAGTTCCCC-3′ | 103–140 | 55 |
| (R) 5′-CTGCAAGGTCTGTTTAACAG-3′ | |||
| D3S1255 | (F) 5′-CTCACTCATGAACACAGATGC-3′ | 145–160 | 55 |
| (R) 5′-AACCCATCTTGTATTCTTGCAG-3′ | |||
| Bat 25 | (F) 5′-TCGCCTCCA AGAATGTAAGT-3′ | ±90 | 55 |
| (R) 5′-TCTGCATTTTAACTATGGCTC 3′ | |||
| Bat 26 | (F) 5′-TGACTACTTTTGACTTCAGCC-3′ | 130–160 | 55 |
| (R) 5′-AACCATTCAACATTTTTAACCC-3′ | |||
| Bat 40 | (F) 5′-ACAACCCTGCTTTTGTTCCT-3′ | 130–160 | 55 |
| (R) 5′-GTAGAGCAAGACCACCTTG-3′ |
F, forward; R, reverse.
PCR reaction
PCR was carried out in 200 μl thin walled PCR tubes. The PCR core kit (Roche Diagnostics) was used for this procedure. The kit consisted of: 10× reaction buffer (containing 1.5mM MgCl2), dNTP mix, and Taq DNA polymerase. The CY5 labelled primers (10 pmoles) were used in the PCR in a total reaction volume of 25 μl, containing 5 μl template DNA, 200μM dNTPs, 50mM PCR buffer containing 1.5mM MgCl2, and 0.75 U Taq DNA polymerase. The PCR amplification was performed using a Techne Progene Thermocycler. The PCR reaction mixture was initially denatured at 95°C for five minutes. Thirty five cycles were performed, consisting of 30 seconds at 94°C, 30 seconds at 55°C, and 40 seconds at 72°C. This was followed by a final extension step for 10 minutes at 72°C.
Sample preparation and DNA fragment analysis
The PCR product (3 μl) was added to 3 μl of loading buffer (98% formamide, 5% dextran blue 2000). The samples were heat denatured at 96°C for three minutes before separation on a 6% Longranger sequencing gel (FMC Bioproducts, Rockland, Maine, USA). The electrophoretic running conditions were 1500 V, 60 mA, and 15 W at a constant temperature of 55°C. The raw data were analysed using the Fragment Manager software (Pharmacia Biotech, Uppsala, Sweden). The preparation of the size marker and the assessment of MSI and LOH were carried out as reported previously.15
RESULTS
Table 2 shows the microsatellite data and table 3 outlines the composite clinical and pathological data.
Table 2.
Summary table showing microsatellite data
| Marker | NLOH | H | LOH | MSI | Percentage informativity |
| D2S123 | 27 | 61 | 7 | 5 | 39 |
| D2S1255 | 25 | 52 | 18 | 5 | 48 |
| D3S659 | 43 | 35 | 21 | 1 | 65 |
| Bat 25 | 12 | 85 | 3 | 0 | 15 |
| Bat 26 | 8 | 83 | 7 | 2 | 17 |
| Bat 40 | 8 | 83 | 5 | 4 | 17 |
H, homozygous with no change; LOH, loss of heterozygosity; MSI, microsatellite instability; NLOH, no loss of heterozygosity.
Table 3.
Clinical and pathological features
| Characteristic | |
| Number of patients | 100 |
| Age range (years) | 24–92 |
| Mean age (years) | 56 |
| Males | 53 |
| Females | 45 |
| Number of patients followed up | 67 |
| Follow up range (weeks) | 0–284 |
| Mean follow up (weeks) | 28 |
| Number of patients alive | 50 |
| Number of patients dead | 17 |
| Pathological data | |
| Stage | |
| I | 1 |
| IIA | 44 |
| IIB | 10 |
| III | 40 |
| Unknown | 5 |
| Tumour grade | |
| Well differentiated | 26 |
| Moderately differentiated | 59 |
| Poorly differentiated | 12 |
| Unknown | 3 |
| Lymph node metastases | |
| No | 41 |
| Yes | 47 |
| No record | 12 |
Molecular analysis
MSI of D2S123 was found in five cases (5%) and LOH was seen in seven of 39 cases. For the D3S659 locus, MSI (fig 1A) was found in only one case (1%), whereas LOH was seen in 21 of the 65 informative cases. MSI of the D3S1255 locus was seen in five cases (5%) and LOH in 18 of 48 informative cases. For the Bat loci (Bat 25, Bat 26, and Bat 40), MSI ranged from 0% to 4%, whereas LOH (fig 1B) ranged from 20% to 41% in the informative cases.
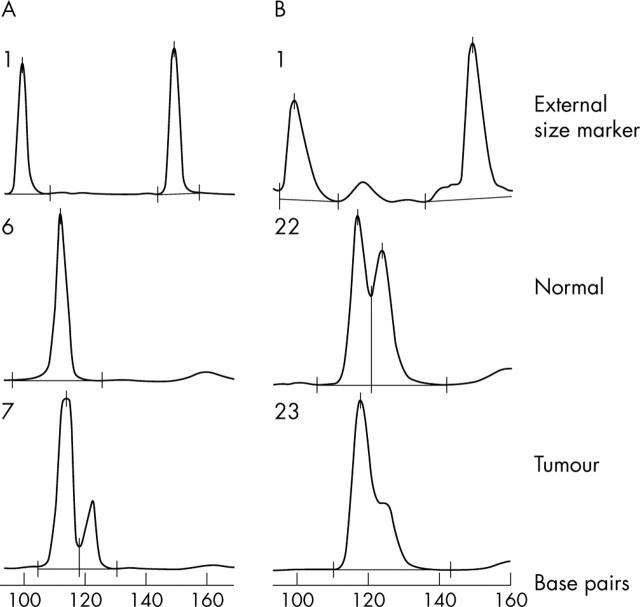
Figure 1.
Representative electropherogram showing microsatellite instability (MSI) and loss of heterozygosity (LOH). (A) MSI for marker D3S659. The normal DNA (lane 6) shows a single peak (homozygous, non-informative case), whereas tumour DNA (lane 7) contains a novel peak (122.6 bp). (B) Electropherogram showing LOH for Bat 25: the middle trace (lane 22) shows normal DNA and the lower trace (lane 23) shows tumour DNA with loss of the larger allele. Lane 1 represents the external marker that was run with all samples.
DISCUSSION
Several risk factors have been identified that are thought to influence the pathogenesis of oesophageal cancer. Among these are nutritional deficiencies, cigarette smoking, excessive alcohol consumption, and exposure to environmental toxins. However, the role of genetic factors in the development of this cancer in this high incidence area have only recently been investigated.7,18–21
MSI is a reflection of the mismatch repair gene status, which is dependent on mutations in human mismatch repair genes hMLH, hPMS1, hPMS2, hMSH2, hMSH3, and hMSH6.13,14 Very few reports have looked at the role of the mismatch repair genes in oesophageal cancer, although several studies have investigated MSI using a range of microsatellite markers located within tumour suppressor genes and oncogenes. Because there are no clear guidelines for the investigation of MSI in oesophageal cancer we adopted the criteria suggested by Boland et al for our study.17 A recent study showed 80% LOH within the TP53 gene in oesophageal cancer.22 Kagawa and co-workers23 investigating MSI in 41 resected oesophageal carcinomas showed that the incidence of MSI was 42%, which was substantially lower than that reported by Wang et al.5 Another study investigating DNA replication error in 30 oesophageal squamous cell carcinomas found that LOH was very high (73%). That study was carried out at seven microsatellite markers in the 2p, 3p, and 16q loci in a cohort of patients from the Indian population.24 Thus, there are large discrepancies in the findings from these studies, probably as a result of the different technologies used in the various studies, the number of cases and microsatellite markers investigated, and also the specific microsatellite markers used in the studies. Another important factor that cannot be ignored is the fact that these findings are a true reflection of the population genetics of the population group being investigated.
“Our results suggest that the molecular pathway for the development of oesophageal cancer is different to mismatch repair gene deficient colorectal cancers”
We found that MSI occurred at a much lower level than that reported in other studies. In contrast, LOH was much higher than MSI and was similar to other published reports, particularly at the 3p region, which showed LOH in 40% of cases. These observations in the 3p region are similar to previous studies,5,25–27 although two other studies28,29 found LOH in the 3p region to be just 10% and 9%, respectively. LOH at the 3p loci was frequently associated with carcinomas that had spread to lymph nodes.26 We found that MSI in the mismatch repair loci ranged from 0% to 5%. These figures are much lower than previous reports.
Take home messages.
We found a low frequency of genetic aberrations in six microsatellite markers located on mismatch repair loci in squamous carcinomas of the oesophagus from a high incidence area in South Africa
No genetic marker at these mismatch repair gene loci predisposed to the onset of oesophageal cancer in our population group
Loss of heterozygosity was much more frequent at these loci and the 3p region seemed to be an important “hot spot” with regard to abnormalities in oesophageal cancer, although this requires further investigation because of the low informativity rate of these markers in our population group
In conclusion, we found a low rate of MSI at the mismatch repair loci in oesophageal cancer in South Africa, whereas LOH was much more frequent at these loci. Furthermore, the 3p region seems to be an important “hot spot” with regard to abnormalities in oesophageal cancer, although this requires further investigation because of the low informativity rate of these markers in our population group. This variation in the informativity rate may result from population genetics. The Bat regions showed minimal instability and are markers of low informativity in our population group. Because of the low informativity of the Bat markers, no clear conclusions can be made with regard to the LOH data at these microsatellite loci.
Our results clearly indicate that there is no genetic marker at these mismatch repair gene loci that predisposes to the onset of oesophageal cancer in our population group. A combination of environmental factors, and/or other genetic factors may have a stronger influence on the onset of the disease. Furthermore, our results suggest that the molecular pathway for the development of oesophageal cancer is different to mismatch repair gene deficient colorectal cancers.
Acknowledgments
This work was funded by grants from the South African Medical Research Council, Cancer Association of South Africa, and the University of Natal Research Fund.
Abbreviations
LOH, loss of heterozygosity
MSI, microsatellite instability
PCR, polymerase chain reaction
REFERENCES
- 1.Sitas F, Madhoo J, Wessie J. Incidence of histologically diagnosed cancer in South Africa, 1993–1995. In: National cancer registry of South Africa. Johannesburg: South African Institute of Medical Research 1998.
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67. [DOI] [PubMed] [Google Scholar]
- 3.Duesberg P, Li R. Multistep carcinogenesis: a chain reaction of aneuploidizations. Cell Cycle 2003;3:202–10. [PubMed] [Google Scholar]
- 4.Jones PA. Epigenetics in carcinogenesis and cancer prevention. Ann N Y Acad Sci 2003;983:213–19. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Li W, Wang X, et al. Genetic alterations on chromosome 6 and 9 of esophageal cancer tissue from China. Oncogene 1996;13:699–703. [PubMed] [Google Scholar]
- 6.Muzeau F, Flejou J, Belghiti J, et al. Infrequent microsatellite instability in oesophageal cancers. Br J Cancer 1997;75:1336–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naidoo R, Tarin M, Reddi A, et al. Allelic imbalance and microsatellite instability in chromosomes 2p, 3p, 5q and 18q in oesophageal squamous carcinoma from South Africa. Diagn Mol Pathol 1999;8:131–7. [DOI] [PubMed] [Google Scholar]
- 8.Ikeguchi M, Unate H, Maeta M, et al. Detection of loss of heterozygosity at microsatellite loci in esophageal squamous-cell carcinoma. Oncology 1999;56:64–8. [DOI] [PubMed] [Google Scholar]
- 9.Kagawa Y, Yoshida K, Hirai T, et al. Microsatellite instability in squamous cell carcinomas and dysplasias of the oesophagus. Anticancer Res 2000;20:213–18. [PubMed] [Google Scholar]
- 10.Mimori K, Inoue H, Shiraishi T, et al. Microsatellite instability is often observed in esophageal carcinoma patients with allelic loss in the FHIT/FRA3B locus. Oncology 2003;64:275–9. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y, Boynton R, Blount PL, et al. Loss of heterozygosity involves multiple tumour suppressor genes in human esophageal cancers. Cancer Res 1992;52:6525–30. [PubMed] [Google Scholar]
- 12.Peltomaki P . DNA mismatch repair and cancer. Mutat Res 2001;488:77–85. [DOI] [PubMed] [Google Scholar]
- 13.Muller A, Fishel R. Mismatch repair and the hereditary non-polyposis colorectal cancer syndrome (HNPCC). Cancer Invest 2002;20:102–9. [DOI] [PubMed] [Google Scholar]
- 14.Peltomaki P . Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol 2003;21:1174–9. [DOI] [PubMed] [Google Scholar]
- 15.Naidoo R, Tarin M, Chetty R. A comparative microsatellite analysis of colorectal cancer in patients under 35 and over 50 years of age. Am J Gastroenterol 2000;95:3266–75. [DOI] [PubMed] [Google Scholar]
- 16.Olansky L, Janssen R, Willing C, et al. Variability of the insulin gene in American blacks with NIDDM. Diabetes 1992;41:742–9. [DOI] [PubMed] [Google Scholar]
- 17.Boland CR, Thibideau SN, Hamilton SR, et al. National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248–59. [PubMed] [Google Scholar]
- 18.Chetty R, Simelane S. p53 and cyclin A protein expression in squamous carcinoma of the oesophagus. Pathol Oncol Res 1999;5:193–6. [DOI] [PubMed] [Google Scholar]
- 19.Gamieldien W, Victor TC, Mugwanya D, et al. p53 and p16/CDKN2 gene mutations in esophageal tumours from a high-incidence area in South Africa. Int J Cancer 1998;78:544–9. [DOI] [PubMed] [Google Scholar]
- 20.Du Plessis L, Dietzsch E, Van Gele M, et al. Mapping of novel regions of DNA gain and loss by comparative genomic hybridization in esophageal carcinoma in the black and colored populations of South Africa. Cancer Res 1999;59:1877–83. [PubMed] [Google Scholar]
- 21.Vos M, Adams CH, Victor TC, et al. Polymorphisms and mutations found in the regions flanking exons 5 to 8 of the TP53 gene in a population at high risk for esophageal cancer in South Africa. Cancer Genet Cytogenet 2003;140:23–30. [DOI] [PubMed] [Google Scholar]
- 22.Hu N, Huang J, Emmert-Buck MR, et al. Frequent inactivation of the TP53 gene in esophageal squamous cell carcinoma from a high-risk population in China. Clin Cancer Res 2001;7:883–9. [PubMed] [Google Scholar]
- 23.Kagawa Y, Yoshida K, Hirai T, et al. Microsatellite instability in squamous cell carcinomas and dysplasias of the esophagus. Anticancer Res 2000;20:213–17. [PubMed] [Google Scholar]
- 24.Mathew R, Arora S, Mathur M, et al. Esophageal squamous cell carcinomas with DNA replication errors (RER+) are associated with p16/pRb loss and wild-type p53. J Cancer Res Clin Oncol 2001;127:603–12. [DOI] [PubMed] [Google Scholar]
- 25.Aoki T, Mori T, Du X, et al. Allelotype study of esophageal carcinoma. Genes Chromosomes Cancer 1994;10:177–82. [DOI] [PubMed] [Google Scholar]
- 26.Ogasawara S, Maesawa C, Tamura G, et al. Frequent microsatellite alterations on chromosome 3p in esophageal squamous cell carcinoma. Cancer Res 1995;55:891–4. [PubMed] [Google Scholar]
- 27.Shibagaki I, Shimada Y, Wagata T, et al. Allelotype analysis of esophageal squamous cell carcinoma. Cancer Res 1994;54:2996–3000. [PubMed] [Google Scholar]
- 28.Wagata T, Ishizaki K, Imamura M, et al. Deletion of 17p and amplification of int-2 gene in esophageal carcinomas. Cancer Res 1991;51:2113–17. [PubMed] [Google Scholar]
- 29.Nakashima H, Mori M, Mimori K, et al. Microsatellite instability in Japanese esophageal carcinoma. Int J Cancer 1995;64:286–9. [DOI] [PubMed] [Google Scholar]