Abstract
AP-2 adaptors regulate clathrin-bud formation at the cell surface by recruiting clathrin trimers to the plasma membrane and by selecting certain membrane proteins for inclusion within the developing clathrin-coat structure. These functions are performed by discrete subunits of the adaptor heterotetramer. The carboxyl-terminal appendage of the AP-2 α subunit appears to regulate the translocation of several endocytic accessory proteins to the bud site. We have determined the crystal structure of the α appendage at 1.4-Å resolution by multiwavelength anomalous diffraction phasing. It is composed of two distinct structural modules, a β-sandwich domain and a mixed α–β platform domain. Structure-based mutagenesis shows that alterations to the molecular surface of a highly conserved region on the platform domain differentially affect associations of the appendage with amphiphysin, eps15, epsin, and AP180, revealing a common protein-binding interface.
Eukaryotic cells take up extracellular macromolecules within small invaginations of the cell surface in a process termed endocytosis. The bulk of endocytosis is believed to occur at discrete bud sites on the plasma membrane coated with a polygonal clathrin lattice. AP-2 adaptors play a pivotal role in the assembly of these clathrin-coated buds (1, 2). The heterotetrameric adaptor, composed of ≈100-kDa α- and β2-, 50-kDa μ2- and 17-kDa σ2 subunits (Fig. 1A), recruits clathrin trimers to the membrane surface, orchestrating the assembly of the clathrin lattice. AP-2 also plays a direct role in selecting molecules for preferential inclusion within the developing clathrin-coated bud (1). The sorting function appears to be mediated primarily by the μ2 subunit of the adaptor complex. μ2 interacts directly, albeit weakly, with tyrosine-based internalization signals present in many transmembrane proteins (3). The μ2 subunit has also been reported to interact directly with the dileucine class of sorting signals (4). The crystal structures of the carboxyl-terminal portion of the μ2 chain associated with peptides containing tyrosine-based internalization signals show that these sorting signals interact with μ2 in an extended conformation (5). Because AP-2 maintains direct interactions with both the overlying clathrin lattice and the sorting signals projecting up from the membrane below, the adaptor is incorporated into clathrin-coated vesicles near stoichiometrically with clathrin.
Figure 1.
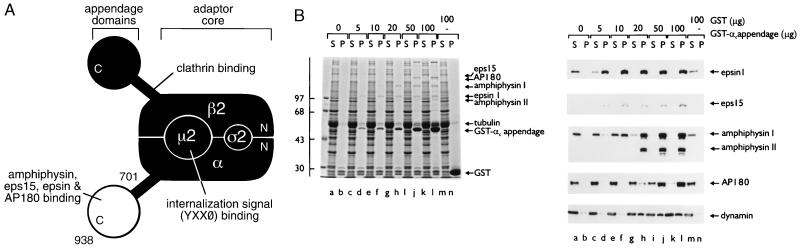
(A) Schematic illustration of the subunit organization of the AP-2 adaptor heterotetramer. Regions of the complex with known protein-binding functions are indicated. For tyrosine-based internalization signals, X is any amino acid and Ø represents a bulky hydrophobic residue (1, 3). (B) Functional protein associations with the GST-αC-appendage fusion protein. Purified GST-αC appendage (0–100 μg) or GST (100 μg), immobilized on 25 μl packed glutathione Sepharose, were incubated in ≈7.5 mg/ml rat brain cytosol for 60 min at 4°C. The Sepharose beads were then recovered by centrifugation, and aliquots corresponding to ≈1/150 of the supernatant (S) and 1/10 of each pellet (P) were resolved by SDS/PAGE and either stained with Coomassie blue (Left) or transferred to nitrocellulose (Right). Portions of the blots were probed with anti-epsin, anti-eps15, anti-amphiphysin, anti-AP180, or anti-dynamin antibodies. The position of the markers (kDa) is indicated on the left and only the relevant portion of each blot is shown.
It is now clear that additional proteins also contribute to the productive assembly of clathrin-coated vesicles. For example, the GTP-binding protein dynamin plays an important role in the final scission process (6). Dynamin appears to be recruited into the assembling lattice by amphiphysin; a proline-based sequence (PSRPNR), located at the carboxyl-terminal end of dynamin, binds to the Src homology 3 (SH3) domain of amphiphysin. The crystal structure of the amphiphysin II SH3 domain reveals that good specificity is achieved for these two binding partners by alteration of the general SH3 fold in amphiphysin to accommodate the uncommon two arginine residues in the dynamin sequence (7). Overexpression of the SH3 domain of amphiphysin inhibits clathrin-mediated endocytosis in vivo by interceding in the translocation of soluble dynamin onto the membrane (8, 9). The inhibition indicates that the recruitment of amphiphysin precedes dynamin and, accordingly, an AP-2-binding element is found within the central region of amphiphysin I (8–13) that allows the protein to bind to the carboxyl-terminal end of the α subunit of AP-2.
Electron microscopic images of purified AP-2 heterotetramers reveal that two separate globular appendages, which correspond to carboxyl-terminal portions of the large α and β2 chains, project off the central core of the complex (14) (Fig. 1A). Like amphiphysin, several other proteins implicated in clathrin-mediated endocytosis also interact with the autonomous α-subunit appendage. Both eps15 (15, 16) and epsin (17) appear to bind to a similar site on the α appendage and, like the amphiphysin SH3 domain, overexpression of the AP-2-binding portion of either protein potently blocks endocytosis in vivo (17, 18). AP180, another clathrin-coat associated protein (19), also binds to the α appendage of AP-2, as does dynamin (10). Although the precise functions of these accessory proteins remain to be delineated, these studies do reveal that the α appendage is a central coordinator of many of the protein–protein interactions that appear necessary for the biogenesis of clathrin-coated vesicles. Here, we present the x-ray crystal structure of the αC-subunit appendage solved at 1.4-Å resolution. The appendage is composed of two linked structural modules, a β-sandwich domain and mixed α–β platform domain. Structure-based mutagenesis reveals that a highly conserved surface on the platform domain of the appendage is a major interface for the association with amphiphysin, epsin, eps15, and AP180.
MATERIALS AND METHODS
Protein Purification and Crystallization.
A construct of the appendage domain (residues 701–938) of the murine αC subunit [the ubiquitously expressed form of the α subunit (20)] in pGEX-2T was kindly provided by R. G. W. Anderson (10). The glutathione S-transferase (GST)-fusion protein was expressed in Escherichia coli BL21 (DE3) by using a standard induction protocol that entails shifting log-phase cultures (A600 = ≈0.6) from 37°C to room temperature for induction. Bacteria were recovered by centrifugation and then the GST-fusion proteins collected on glutathione Sepharose 4B after lysis of the bacteria in B-PER reagent (Pierce) and removal of insoluble material by centrifugation. After washing the beads extensively with PBS, the αC appendage was released by thrombin cleavage, exchanged into 25 mM Tris⋅HCl, pH 7.5/25 mM NaCl, and further purified by sequential anion-exchange and gel-filtration chromatography. The purified appendage was concentrated to ≈6 mg/ml in a Centricon (Amicon) device for crystallization. After thrombin cleavage, the linker sequence GSPGIRLGS precedes Ser-701 of the αC appendage. Selenomethionine (SeMet)-substituted αC appendage was prepared after transforming the pGEX construct into the E. coli strain DL41 (21). A starter culture (100 ml), in modified LeMasters medium (22) containing 25 μg/ml l-SeMet, was used to inoculate the large-scale (4-liter) culture in the same medium. Induction and purification was as described above for the native protein except that all buffers contained 5 mM EDTA and 10 mM DTT. Mass spectroscopy showed that the replacement of the four Met positions by SeMet was quantitative. Crystallization of the purified αC-appendage was achieved by hanging drop vapor diffusion by using a wide array of crystallization conditions. The best native crystals were grown from 1.3 M ammonium sulfate/80 mM Hepes, pH 6.9/8% dioxane at 20°C. Drops were set up by using 1 μl of protein at 6 mg/ml mixed with 0.5 μl of precipitant buffer. These crystals can grow as large as 250 μm × 250 μm × 75 μm and belong to the monoclinic space group P21. The unit cell dimensions are a = 40.81 Å, b = 72.75 Å, c = 41.86 Å, b = 99.69°. The best diffracting crystals of the SeMet-substituted αC appendage were grown from 1.5 M magnesium sulfate and 100 mM Mes, pH 6.8, at 20°C. Drops were set up with 1.3 μl of protein at 9 mg/ml with 0.6 μl well solution and 0.1 μl 100 mM spermine tetra-HCl. These crystals are morphologically indistinguishable from the P21 crystals but belong to the triclinic space group P1 with unit cell dimensions a = 38.70 Å, b = 40.73 Å, c = 41.84 Å, α = 99.68°, β = 95.81°, γ = 113.60°. Both crystal forms were cryo-preserved by liquid nitrogen flash-cooling by using 30% glycerol in combination with the crystal mother liquor.
Phase Determination and Structure Refinement.
The structure of the αC appendage was determined by the multiwavelength anomalous diffraction (MAD) phasing method (23). Four data sets were collected for the SeMet protein in the P1 crystal form at wavelengths near the Se absorption edge at the Advanced Photon Source (APS) undulator beamline 19-ID on a charge-coupled device detector (24). Data were indexed and processed by using denzo and scalepack (25). Atomic positions for the four Se atoms were obtained by means of Patterson methods with initial positions refined by using mlphare (CCP4) (26). MAD phasing was carried out by using the program sharp (27). The resulting experimental map was of very high quality, with an overall figure of merit of 0.67 to 1.6 Å (0.94 after solvent flattening/flipping by using solomon (CCP4)). The atomic model was traced with the program o (28). The model was then refined with the programs x-plor (29) and refmac (CCP4). Data for the native crystal in the P21 space group were also collected at APS 19-ID. The structure was solved by molecular replacement by using the P1 crystal form coordinates with the program amore (30). For both the P1 and P21 structures, there were no ambiguous regions such that complete atomic models could be accurately built and refined for all 238 residues of the αC appendage as well as the amino-terminal nine residues corresponding to the fusion protein linker region.
Binding Assays.
The association of cytosolic proteins with the GST-αC appendage was assayed in 25 mM Hepes-OH, pH 7.2/125 mM potassium acetate/2.5 mM magnesium acetate/5 mM EGTA/1 mM DTT (assay buffer) in a final volume of 500 μl. GST or the GST-fusion proteins were first each immobilized on 25 μl packed glutathione Sepharose, and the immobilized proteins were then washed and resuspended to 100 μl in assay buffer. Rat brain cytosol (31) was added and the tubes incubated at 4°C for 60 min with constant gentle mixing. The Sepharose beads were then recovered by centrifugation at 10,000 × gmax for 1 min and 35-μl aliquots of each supernatant removed and adjusted to 100 μl with SDS/PAGE sample buffer. After washing the Sepharose pellets four times each with ≈1.5 ml ice-cold PBS by centrifugation, the supernatants were aspirated and each pellet resuspended to a volume of 100 μl in SDS/PAGE sample buffer. Unless otherwise indicated, 10-μl aliquots, equivalent to ≈1/150 of each supernatant and 1/10 of each pellet, were resolved by SDS/PAGE and either stained with Coomassie blue or transferred to nitrocellulose. Note that decreased crosslinking as a result of the altered acrylamide:bis-acrylamide (30:0.4) ratio we use for SDS/PAGE affects the mobility of several proteins, most noticeably eps15, AP180, and epsin. Blots were probed with polyclonal antibodies against epsin (17) or with monoclonal antibodies against amphiphysin I and II, AP180, dynamin, and eps15 (Transduction Laboratories, Lexington, KY). Bound antibodies were visualized by using enhanced chemiluminescence.
Mutagenesis.
αC appendage mutants were generated by site-specific mutagenesis with the QuikChange Kit (Stratagene) by using the GST-αC in pGEX-2T as the template. The sense primers used to generate the F837A, R905A, and R916A mutants were 5′-GGCTTCTCAGGATTTTGCGCAACGTTGGAAGCAGTTGAGC-3′, 5′-CCCAGATTGGCTGCTTGTTGGCGCTGGAGCCAAACC-3′, and 5′-CCTGCAAGCTCAGATGTACGCGTTGACCTTGCGTACCAGC-3′, respectively. All mutants were verified by automated dideoxy sequencing.
RESULTS AND DISCUSSION
The general function of the AP-2 adaptor complex is to recruit select proteins to a clathrin-bud site. The appendage of the α subunit binds to several endocytic accessory proteins and, when expressed as a GST-fusion protein, the αC appendage associates with soluble accessory proteins present in brain cytosol in a hierarchical fashion (Fig. 1B). In our assays, epsin 1 and eps15 display the highest apparent affinity for the functional appendage domain. Even at very low concentrations of immobilized GST-αC, both proteins translocate near quantitatively from the cytosol fraction onto the beads (Fig. 1B). Amphiphysin I and II and AP180 also move almost completely onto the beads, but both require higher concentrations of the immobilized appendage to do so. Dynamin interacts with the αC appendage only poorly and, unlike the other endocytic accessory proteins, the bulk of the dynamin input remains in the soluble fraction (Fig. 1). Because dynamin does not appear to associate extensively with the GST-αC in the absence of amphiphysin, our data support the idea that dynamin most likely associates with AP-2 predominantly by binding to the SH3 domain of amphiphysin (7).
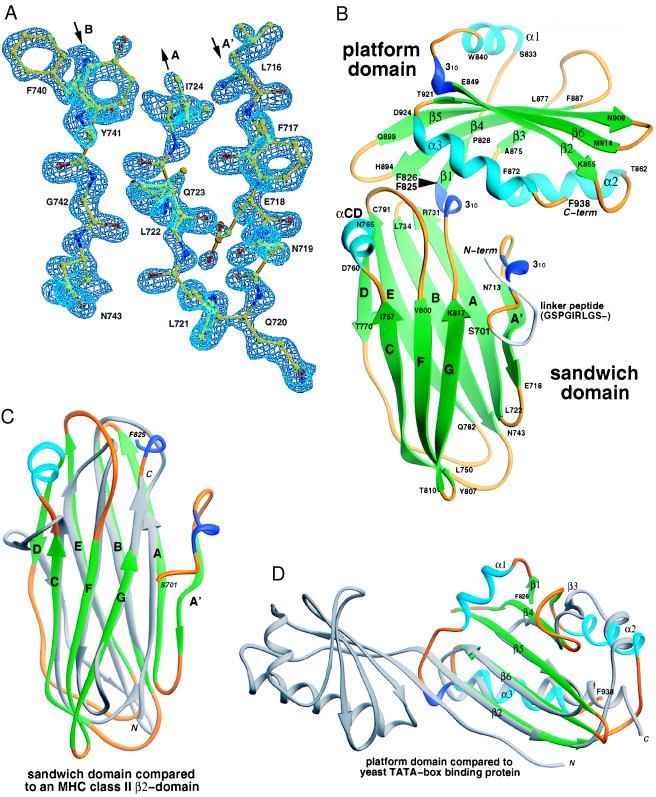
Because the primary sequence of the αC appendage does not provide any obvious clues as to how several accessory proteins can bind to this domain, we have solved the x-ray crystal structure of this biologically active fragment. The structure of the SeMet-substituted protein was determined at 1.6-Å resolution by MAD phasing (23) (Fig. 2A, Table 1), and the structure of the native protein was refined in a second space group at 1.4-Å resolution (Table 2). The αC appendage is a two-lobed structure of approximate dimensions 47 Å × 53 Å × 59 Å (Fig. 2B). The first domain (residues Ser-701–Phe-825), proximal to the AP-2 adaptor core, is a two-sheet β-sandwich composed of eight antiparallel β-strands. The distal domain (residues Phe-826–Phe-938) is a mixed α-β-folded structure we term the platform domain, composed of two helices buttressing an antiparallel β-sheet and with an additional helix crossing the top of the sheet. The two domains make extensive interactions with each other (>1,200 Å2 of buried surface area), and no segmental mobility is observed in comparing the structures in two different crystal-packing environments (overall rms deviation of 0.35 Å).
Figure 2.
The AP-2 αC-appendage structure. (A) Experimental electron-density map of the α appendage. A portion of the MAD-phased electron density at 1.6-Å resolution for the P1 crystal form is shown with the refined atomic model superimposed. (B) Ribbon representation (32) of the αC appendage. α-helices are colored in cyan, β-strands in green, 310 helical segments in blue, and connecting loops in gold. The black arrowhead indicates the domain boundary. (C and D) Topological similarities. (C) Superimposition of the appendage-sandwich domain with a IG superfamily constant domain, the class II MHC I-Ak β2 domain (33). A total of 79 residues of the β2 domain can be aligned with 125 residues of the appendage with a rms deviation (rmsd) of 3.8 Å. Only 5% of the residues are identical. Comparison of the β-sandwich domain (34) reveals extensive structural overlap with subdomain B of the μ2 subunit of AP-2 (5), with 100 of 125 residues aligned with an rmsd of 2.8 Å, although the topology is distinct and the sequence identity for aligned residues is only 8%. (D) Superimposition of the appendage-platform domain with TBP (35). TBP is pseudosymmetrical, composed of two structural repeats which, together, form a concave saddle-like surface over which the TATA box runs. Comparison of the platform domain with the TBP carboxyl-terminal repeat aligns 75 of 113 residues with an rmsd of 3.0 Å and sequence identity of 8%. The orientation of the platform domain has been rotated forward relative to B.
Table 1.
Summary of crystallographic analysis
| Diffraction data | PI SeMEt-1 | P1 SeMet-2 | P1 SeMet-3 | P1 SeMet-4 | P21-Native | |
|---|---|---|---|---|---|---|
| Wavelength, Å | 1.07813 | 0.97956 | 0.97945 | 0.94645 | 0.97945 | |
| Resolution, Å (outer shell) | 100–1.60 (1.66–1.60) | 100–1.60 (1.66–1.60) | 100–1.60 (1.66–1.60) | 100–1.60 (1.66–1.60) | 100–1.40 (1.45–1.4) | |
| Number of sites | 4 | 4 | 4 | 4 | — | |
| Reflections | Measured | 126,734 | 146,273 | 146,405 | 149,455 | 495,848 |
| (unique) | (49,265) | (56,587) | (56,619) | (57,749) | (45,711) | |
| Completeness | Overall | 81.8 | 93.9 | 94.0 | 95.8 | 96.1 |
| (outer shell) | (35.3) | (80.4) | (80.9) | (94.8) | (82.1) | |
| I/σ(I) | Overall | 22.5 | 26.8 | 21.9 | 19.1 | 23.0 |
| (outer shell) | (7.9) | (11.6) | (9.8) | (7.3) | (3.0) | |
| Rsym, % | Overall | 5.9 | 5.8 | 6.0 | 6.5 | 10.0 |
| (outer shell) | (19.2) | (17.0) | (19.3) | (28.6) | (67.2) | |
| Rcullis, 20–1.6 Å | iso/ano | —/0.967 | 0.359/0.563 | 0.283/0.529 | 0.607/0.729 | — |
| Phasing power | iso/ano | —/0.818 | 4.56/3.10 | 6.19/3.30 | 0.819/2.31 | — |
Table 2.
Refinement statistics
| Refinement | P1 SeMet | P21 Native |
|---|---|---|
| Resolution range, Å | 20.0–1.60 (1.66–1.60) | 20.0–1.40 (1.45–1.40) |
| Number of reflections/ completion | 29,053/97.1% (2,750/95.7%) | 45,632/96.3% (3,965/84.6%) |
| Number of protein atoms/solvent atoms | 1,957/266 | 1,957/244 |
| Rcrystal/Rfree, % | 16.8/21.1 (26.2/27.8) | 17.8/20.9 (26.0/26.3) |
| rms deviations | ||
| Bonds, Å | 0.010 | 0.011 |
| Angles, ° | 1.9 | 1.5 |
A search for structural similarity to the αC appendage found no similar two-lobed structures, although both individual domains do have structural precedents. The β-sandwich domain has a topology remarkably similar to proteins of the Ig superfamily (IGSF), with a single additional β-strand (termed A′) relative to a canonical IGSF C-type fold (Fig. 2C). The Cys residues that traditionally form a disulfide bond between the B and F strands of IGSF domains are replaced by hydrophobic residues in the αC appendage (Ile-739 and Leu-801). We suspect that the presence of this topology in the appendage might be another example of evolutionary convergence on a highly stable fold (36). The structure of the platform domain displays considerable topological similarity (34) with the yeast TATA-box binding protein (TBP) (35). The major differences between the TBP fold and the αC appendage platform domain are the lack of an overlying α-helix and the addition of a single β-strand at the edge of the sheet in TBP (Fig. 2D).
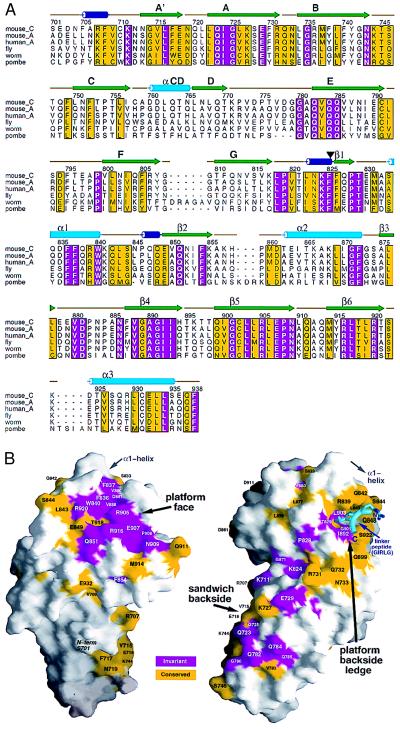
Sequence alignment of the appendage domain of mouse αC with those of α subunits from a variety of species reveals a substantial number of invariant residues (Fig. 3A). Primary sequence identity is greatest in the platform domain, where almost 25% of the residues (28 of 113 in mouse αC) are invariant. The distribution of the conserved residues on the three-dimensional surface of αC reveals three general areas of invariance: the platform face, a ledge on the backside of the platform, and the backside of the sandwich domain below the ledge (Fig. 3B). The conserved surface on the backside of the sandwich domain is somewhat discontinuous, and the majority of residues are hydrophilic. The backside ledge below the platform domain α1-helix, on the other hand, has a hydrophobic character. Interestingly, a region of the 9-aa linker peptide binds deeply into the hydrophobic ledge of a crystallographically related appendage in both the P1 and P21 crystal forms. A number of invariant and conserved α-appendage residues are contacted by two hydrophobic residues of the linker peptide, suggesting a possible role for this region in mediating interactions with one or more binding partner. Far more striking, however, is the absolute invariance of the surface of the platform face (Fig. 3B). Remarkably, half of the invariant residues in the platform domain converge in this area, strongly suggesting a highly conserved function. In the middle of the conserved patch is a hydrophobic pocket created in part by three residues from the sheet-crossing α1-helix (Phe-836, Phe-837, and Trp-840) and four residues coming from other secondary structures (Val-880, Val-888, and the aliphatic portions of Asp-881 and Arg-905) (Figs. 3B and 4B). The hydrophobic character of the solvent-exposed residues in this cluster raises the possibility that this surface interacts with a binding ligand (37). Interestingly, the topological alignments (Fig. 2) show that the molecular surface of TBP that interacts with DNA (35) overlaps the clustered patch of conserved residues in the αC-platform domain (Fig. 2). The overlap in the binding surfaces suggests that the fold might facilitate macromolecular associations in general and supports the idea that the conserved region of the αC platform is an essential surface for the functioning of the appendage.
Figure 3.
Sequence conservation in α-subunit appendages. (A) clustalx (38) alignment of the appendage domains of Mus musculus αC (mouse_C), M. musculus αA (mouse_A), Homo sapiens αA (human_A), and Drosophila melanogaster (fly), Caenorhabditis elegans (worm), and Schizosaccharomyces pombe (pombe) α subunits. Residue numbers above the sequences correspond to mouse αC sequence. Residues shaded in magenta are invariant, whereas yellow-colored residues have a conservation index of 7 or greater, as determined by the program alscript (39). The secondary structures are displayed and colored as in Fig. 2 with the domain boundary between Phe-25 and Phe-826 demarcated by an arrow. (B) Mapping of residue conservation onto the surface of the αC appendage (40), colored as in A. The orientation of the view on the left has been rotated forward relative to Fig. 2B to expose the platform face. The protein has been rotated approximately 180° on the right to view the backside of the appendage. This view also shows the location of the linker peptide (a postthrombin cleavage artifact, colored cyan), derived from a crystallographically related appendage in two different space groups, bound to the platform backside ledge. The largest contiguous invariant surface is clearly the platform face.
Figure 4.
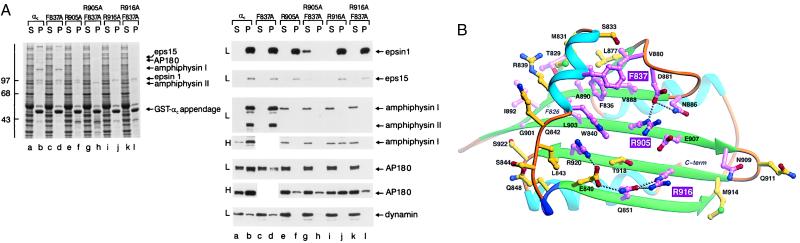
αC appendage–partner recognition surface. (A) Glutathione Sepharose (25 μl packed beads) containing either wild-type GST-αC appendage (αC) or GST-αC appendage F837A, R905A, R905A-F837A, R916A, or R916A-F837A mutants were incubated with rat brain cytosol for 60 min at 4°C. The Sepharose beads were then recovered by centrifugation, and aliquots corresponding to 1/150 of the supernatant (S) and 1/10 of each washed pellet (P) were resolved by SDS/PAGE and either stained with Coomassie blue (Left) or transferred to nitrocellulose (Right). Portions of the blots were probed with anti-epsin, anti-eps15, anti-amphiphysin, anti-AP180, or anti-dynamin antibodies. Immunoblots from assays performed at low (≈25 μg, L) or high (≈100 μg, H) GST-αC density are indicated on the left. In the experiment performed at high density shown, the GST-αC F837A mutant was not tested, and 1/40 of each supernatant was analyzed. The position of the markers (kDa) is indicated on the left, and only the relevant portion of each blot is shown. (B) Ribbon diagram (32) of the platform domain viewed from the top. Conserved residues that make up the upper surface of the platform are colored with invariant residues shaded magenta and conserved residues, yellow. The extended hydrogen-bonding network is shown as small gray balls with oxygen atoms in red, nitrogen in blue. The three highly exposed residues that have major consequences on partner binding when mutated to Ala are highlighted (F837, R905, R916).
To test experimentally whether the platform surface is required for binding to endocytic accessory proteins, several appendage mutants were prepared, and each was allowed to associate with the accessory proteins present in brain cytosol. The effects of the substitutions F837A, R905A, and R916A, all invariant surface residues within the conserved cluster (Figs. 3 and 4B), are shown (Fig. 4A). The F837A mutation significantly reduces the interaction of AP180 with the GST-αC appendage, but has little effect on the binding of amphiphysin, epsin, eps15, or dynamin. An R905A substitution has a more dramatic effect, virtually eliminating the association of both amphiphysin I and II and AP180. Although binding of eps15 and epsin to GST-αC is slightly inhibited in the R905A mutant, this substitution, together with F837A, abolishes the ability of both proteins to bind to the αC appendage. Arg-916, located adjacent to Arg-905 (Fig. 4B), also contributes to the binding surface for amphiphysin and AP180, because an R916A change alone strongly inhibits the recruitment of both proteins (Fig. 4A). Because of the different apparent affinities of amphiphysin and AP180 for the αC appendage (Fig. 1), the effect of the F837A and R916A mutations varies with the density of the GST-αC-fusion protein immobilized on the glutathione Sepharose. At low density, the R916A mutant binds neither AP180 nor amphiphysin. At ≈4-fold higher density, detectable binding of both proteins is seen, and association with the appendage is abolished only in the R916A-F837A double mutant. Interestingly, the residues equivalent to Arg-905 (Leu-205) and Arg-916 (Lys-211) in TBP both contact the TATA DNA element directly (35). Arg-916 does not appear to contribute to the binding surface on the platform for either epsin or eps15 as binding to R916A, and the R916A-F837A double mutant is still normal compared with the wild-type protein (Fig. 4A). The weak binding of dynamin to the αC appendage responds to the mutations, in general, in an analogous way to amphiphysin (Fig. 4). However, trace association of dynamin occurs in the R905A-F837A mutant, in the absence of detectable amphiphysin binding, suggesting that dynamin might associate with the appendage directly (10), albeit very weakly.
Taken together, our data show that a surface containing numerous highly conserved residues at the top of the platform domain forms a principal binding interface for several endocytic accessory proteins. Deleting residues 701–750 of the αC appendage results in a protein that is unable to bind either eps15 (15) or epsin 1 (17). Because the appendage-binding carboxyl-terminal portion (residues 501–874) of eps15 is able to compete with the central domain (residues 249–401) of epsin 1 for binding to the αC appendage (17), it has been concluded that an overlapping-binding site for these two proteins lies within residues 701–750 of αC (41). These residues encompass the first three β strands (A′, A, and B) of the sandwich domain (Fig. 2). Removal of this portion of the appendage would severely disrupt the hydrophobic core of the domain and delete several critical side chains that interact with the overlying platform domain, in particular Phe-705, Ala-706, Val-709, Cys-710, Gln-732, and Asn-733. Because we have shown that the principal binding interface is found on the platform domain, we favor the conclusion that, instead of representing a binding site for epsin and eps15, the structural consequences of truncation of the first 50 amino acids of the αC appendage preclude binding of eps15 and epsin at distant sites.
The precise molecular details of the binding interface between the α appendage and accessory proteins should come from cocrystallization studies. Discrete regions of eps15 (15, 16), epsin 1 (17), and amphiphysin I (42) that are responsible for binding to the αC appendage have been identified. In eps15 and epsin 1, the adaptor-binding portions are characterized by multiple repeats of the tripeptide motifs DPF or DPW, respectively, and the DPF-containing portion of eps15 can inhibit the association of the DPW domain of epsin 1 with expressed αC appendage (17). αC mutations affect the binding of eps15 and epsin 1 identically in our assays, fortifying the notion that the DPF/W repeats might interact with the α appendage. In fact, Arg-905 and Phe-837, the residues we found that, when both are altered to Ala, abolish the binding of these two proteins, are located on the perimeter of the hydrophobic pocket (Fig. 4B) that is well suited to accommodate DPW/F. The region of human amphiphysin I that binds to αC has been mapped to residues 322–375 (42) and, strikingly, this region also contains a DPF sequence at the end of a functional clathrin-binding motif 351LLDLDFDPF (43). Rat brain AP180 has two DPF repeats (residues 399–401 and 474–476) as well as several redundant DXF repeats. Remarkably, auxilin, a necessary cofactor in the Hsc70-mediated clathrin-uncoating process (44), also contains a DPF motif (amino acids 579–581) and associates with the αC appendage (E. Ungewickell, personal communication). Taken together, all these observations provide compelling evidence for the direct involvement of the DPF/W sequence in appendage binding and a general explanation for the binding of multiple endocytic accessory proteins to the conserved site on the platform domain. Dynamin 1 and 2 also each contain a single DPF triplet, located six residues ahead of the amphiphysin-binding sequence PSRPNR. Although this may explain the direct association of dynamin with the α appendage (10), it seems likely that the primary mode of association with AP-2 is indirect via amphiphysin (7, 9). Other regions on the molecular surface of the accessory proteins must also be involved in platform recognition to explain their different apparent affinities for the appendage (Fig. 1) and the differential effects of the αC mutations on accessory protein binding (Fig. 4A).
In conclusion, the structure of the α-subunit appendage reveals a molecular platform surface that, as the principal protein-binding interface, is well suited to act as a springboard for coordinating the multiple protein–protein interactions that appear necessary to assemble a clathrin-coated vesicle. Unlike AP-2, amphiphysin, epsin and eps15 are not major components of isolated clathrin-coated vesicles (L.M.T., unpublished results) (17). The association of these proteins with AP-2 appears to be precisely regulated by phosphorylation cascades (41, 42). Our structural description of the binding interface on the α appendage represents a first step toward understanding the molecular basis of this regulation.
Acknowledgments
We are grateful to Richard G. W. Anderson and Pietro De Camilli for generously providing critical materials. We thank Chris Nelson, Yudong Wu, and Robert Latek for aid in synchrotron data collection, Matthew Drake for help with the mutagenesis, and Chris Nelson and Matthew Drake for constructive criticism of the manuscript. This work was supported in part by a Howard Hughes Medical Institute pilot project grant, a Barnes–Jewish Hospital Foundation Grant, National Institutes of Health grant DK53249 (L.M.T.), and in part by the Kilo Diabetes and Vascular Research Foundation (D.H.F.). D.H.F. is the recipient of a Burroughs–Wellcome Fund Career Award in the Biomedical Sciences.
ABBREVIATIONS
- TBP
TATA box-binding protein
- GST
glutathione S-transferase
- MAD
multiwavelength anomalous diffraction
- SeMet
selenomethionine
- SH3
Src homology 3
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1QTP and 1QTS).
A Commentary on this article begins on page 8809.
References
- 1.Kirchhausen T, Bonifacino J S, Riezman H. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- 2.Schmid S L. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- 3.Ohno H, Stewart J, Fournier M-C, Bosshart H, Rhee I, Miyatake S, Saito T, Galluser A, Kirchhausen T, Bonifacino J S. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 4.Bremnes T, Lauvrak V, Lindqvist B, Bakke O. J Biol Chem. 1998;273:8638–8645. doi: 10.1074/jbc.273.15.8638. [DOI] [PubMed] [Google Scholar]
- 5.Owen D J, Evans P R. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmid S L, McNiven M A, De Camilli P. Curr Opin Cell Biol. 1998;10:504–512. doi: 10.1016/s0955-0674(98)80066-5. [DOI] [PubMed] [Google Scholar]
- 7.Owen D J, Wigge P, Vallis Y, Moore J D, Evans P R, McMahon H T. EMBO J. 1998;17:5273–5285. doi: 10.1093/emboj/17.18.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shupliakov O, Low P, Grabs D, Gad H, Chen H, David C, Takei K, De Camilli P, Brodin L. Science. 1997;276:259–263. doi: 10.1126/science.276.5310.259. [DOI] [PubMed] [Google Scholar]
- 9.Wigge P, Vallis Y, McMahon H T. Curr Biol. 1997;7:554–560. doi: 10.1016/s0960-9822(06)00254-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang L H, Sudhof T C, Anderson R G. J Biol Chem. 1995;270:10079–10083. doi: 10.1074/jbc.270.17.10079. [DOI] [PubMed] [Google Scholar]
- 11.David C, McPherson P S, Mundigl O, De Camilli P. Proc Natl Acad Sci USA. 1996;93:331–335. doi: 10.1073/pnas.93.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramjaun A R, Micheva K D, Bouchelet I, McPherson P S. J Biol Chem. 1997;272:16700–16706. doi: 10.1074/jbc.272.26.16700. [DOI] [PubMed] [Google Scholar]
- 13.McMahon H T, Wigge P, Smith C. FEBS Lett. 1997;413:319–322. doi: 10.1016/s0014-5793(97)00928-9. [DOI] [PubMed] [Google Scholar]
- 14.Heuser J E, Keen J. J Cell Biol. 1988;107:877–886. doi: 10.1083/jcb.107.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benmerah A, Begue B, Dautry-Varsat A, Cerf-Bensussan N. J Biol Chem. 1996;271:12111–12116. doi: 10.1074/jbc.271.20.12111. [DOI] [PubMed] [Google Scholar]
- 16.Iannolo G, Salcini A E, Gaidarov I, Goodman O B, Jr, Baulida J, Carpenter G, Pelicci P G, Di Fiore P P, Keen J H. Cancer Res. 1997;57:240–245. [PubMed] [Google Scholar]
- 17.Chen H, Fre S, Slepnev V I, Capua M R, Takei K, Butler M H, Di Fiore P P, De Camilli P. Nature (London) 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- 18.Benmerah A, Lamaze C, Begue B, Schmid S L, Dautry-Varsat A, Cerf-Bensussan N. J Cell Biol. 1998;140:1055–1062. doi: 10.1083/jcb.140.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahle S, Ungewickell E. EMBO J. 1986;5:3143–3149. doi: 10.1002/j.1460-2075.1986.tb04621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ball C L, Hunt S P, Robinson M S. J Cell Sci. 1995;108:2865–2875. doi: 10.1242/jcs.108.8.2865. [DOI] [PubMed] [Google Scholar]
- 21.Hendrickson W A, Horton J R, LeMaster D M. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qoronfleh M W, Ho T F, Brake P G, Banks T M, Pulvino T A, Wahl R C, Eshraghi J, Chowdhury S K, Ciccarelli R B, Jones B N. J Biotechnol. 1995;39:119–128. doi: 10.1016/0168-1656(94)00149-7. [DOI] [PubMed] [Google Scholar]
- 23.Hendrickson W A. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 24.Westbrook E M, Naday I. Methods Enzymol. 1997;276:244–268. [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. In: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S, editors. Warrington, U.K.: SERC Daresbury Laboratory; 1993. pp. 59–62. [Google Scholar]
- 26.Collaborative Computational Project. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 27.de La Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 28.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Brunger A T. x-plor. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1 Manual. [Google Scholar]
- 30.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 31.Traub L M, Bannykh S I, Rodel J E, Aridor M, Balch W E, Kornfeld S. J Cell Biol. 1996;135:1801–1804. doi: 10.1083/jcb.135.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carson M. J Mol Graphics. 1987;5:103–106. [Google Scholar]
- 33.Fremont D H, Monnaie D, Nelson C A, Hendrickson W A, Unanue E R. Immunity. 1998;8:305–317. doi: 10.1016/s1074-7613(00)80536-1. [DOI] [PubMed] [Google Scholar]
- 34.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 35.Kim Y, Geiger J H, Hahn S, Sigler P B. Nature (London) 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 36.Shapiro L, Kwong P D, Fannon A M, Colman D R, Hendrickson W A. Proc Natl Acad Sci USA. 1995;92:6793–6797. doi: 10.1073/pnas.92.15.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young L, Jernigan R L, Covell D G. Protein Sci. 1994;3:717–729. doi: 10.1002/pro.5560030501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barton G J. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 40.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 41.Chen H, Slepnev V I, Di Fiore P P, De Camilli P. J Biol Chem. 1999;274:3257–3260. doi: 10.1074/jbc.274.6.3257. [DOI] [PubMed] [Google Scholar]
- 42.Slepnev V I, Ochoa G C, Butler M H, Grabs D, De Camilli P D. Science. 1998;281:821–824. doi: 10.1126/science.281.5378.821. [DOI] [PubMed] [Google Scholar]
- 43.Ramjaun A R, McPherson P S. J Neurochem. 1998;70:2369–2376. doi: 10.1046/j.1471-4159.1998.70062369.x. [DOI] [PubMed] [Google Scholar]
- 44.Ungewickell E, Ungewickell H, Holstein S E H, Lindner R, Prasad K, Barouch W, Martin B, Greene L E, Eisenberg E. Nature (London) 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]