Abstract
Background: The use in many countries of acid fixatives, such as Bouin’s solution, has limited the use of archival tissue for molecular analysis. An acidic environment is one of the main causes of DNA degradation. Moreover, RNA extraction is difficult in these types of fixed tissues.
Aims: To amplify DNA and RNA from Bouin’s fixed tissues.
Methods: DNA and RNA were extracted from 20 breast cancer samples that had been routinely fixed in Bouin’s fixative. Amplification of several genes using primers that produced amplicons of different lengths was carried out using the polymerase chain reaction (PCR) for DNA (with and without restoration) and reverse transcription PCR for RNA.
Results: The acid environment of Bouin’s fixative damaged both DNA and RNA. However, amplification was successful when the amplicon length was reduced to about 80 bp for RNA and 100–200 bp for DNA, especially if submitted to DNA reconstruction procedures.
Conclusions: It is possible to recover and analyse DNA and RNA from Bouin’s fixed and paraffin wax embedded tissues.
Archival tissue is the most widely available material for retrospective clinical studies. With the potential of molecular analysis of nucleic acids, these tissues represent an invaluable resource for the elucidation of disease mechanisms and the validation of gene expression as prognostic or therapeutic target indicators. The most widely used fixative is neutral formalin, but for special purposes other fixatives can be used in histopathology laboratories. Among them, Bouin’s solution is often used in French and Canadian laboratories for historical reasons. This fixative solution is composed of a saturated aqueous solution of picric acid, formalin, and acetic acid.1 The use of Bouin’s solution is labour intensive, requiring multiple alcohol rinses to remove picric acid for optimal preservation and immunohistochemical detection. Tissues are damaged when the picric acid is not removed properly and remains in the tissue throughout the entire processing. In effect, the harmful reaction continues within the embedded specimen for several years.1 This fixation method is also used for specific purposes because it better conserves some morphological details, such as nuclear conformation. This property makes Bouin’s solution the elective fixative for testis2 and lymphoma specimens.3
There are conflicting reports about the possible use of these specimens for molecular analyses. Many authors have reported the impossibility of analysing DNA extracted from these tissues with the polymerase chain reaction (PCR).4,5 The main reason is that the enhanced acidic environment causes DNA degradation. Moreover, RNA extraction is difficult in these types of fixed tissues.6,7 We have been able to amplify 200 bp fragments of DNA obtained from Bouin’s fixed and paraffin wax embedded tissues only after a specific restoration method to produce longer reconstructed DNA fragments.8
“Bouin’s solution is the elective fixative for testis and lymphoma specimens”
Whereas most reports have denied the possibility of extracting RNA from routinely paraffin wax embedded, Bouin’s fixed specimens,6,9 we have been able to amplify short sequences of RNA/cDNA about 80 bp long.
MATERIALS AND METHODS
Twenty breast cancer samples were obtained from the pathology department of the National Institute for Cancer Study of Milan, Italy, because most of the material in this institution was routinely fixed in Bouin’s solution in the past. All cases chosen were infiltrating ductal breast carcinomas with the same tumour grade and stage, so that histologically homogeneous samples were analysed. All biopsies were carried out from 1980 to 1985, and the fixation and paraffin wax embedding procedure was performed routinely in clinical laboratories. The Bouin’s solution used during that period was traditionally prepared in the laboratory, rather than using a commercially manufactured one. DNA and RNA were extracted from 6 μm thick sections of Bouin’s fixed tissues, as reported previously.10–12 Ten sections were cut for each sample.
DNA amplification
Every DNA sample was restored taking into consideration that the DNA degraded fragments contain mostly single strand nicks, as reported previously.8 DNA samples were briefly incubated for one hour at 55°C in 100 μl of restoration solution containing 100mM Tris/HCl (pH 8.3), 1.5mM MgCl2, 2% Triton X-100, and 200mM of each dNTP. After this step, 1 U of classic Taq DNA polymerase (Amersham Biosciences, Uppsala, Sweden) was added and DNA polymerisation was performed at 72°C for 20 minutes. Treated samples were then stored at −20°C until processed.
Before the PCR was carried out a denaturation step was performed: 10 μl of the solution was denatured at 95°C for five minutes and then immediately chilled on ice. As previously reported,8 the denaturation step is necessary to destroy the secondary structure of reconstructed DNA. For PCR analysis, the sequences of EGFR (epidermal growth factor receptor), p16, and TTR (transthyretin gene) were amplified with the primers described in table 1.
Table 1.
Sequence of primers and PCR conditions for DNA analysis
| Gene | Primer sequence 5′–3′ | Length of product | Ta | Cycles |
| EGFR | Forward: GAA GCC AAG CCA AAT GGC A | 90 bp | 60 | 45 |
| Reverse: GCT CCA ATA AAT TCA CTG CT | ||||
| p16 | Forward: AGC TTC CTT TCC GTC ATG C | 204 bp | 60 | 45 |
| Reverse: 5-GCA GCA CCA CCA GCG TG | ||||
| TTR1 | Forward: CAG CAG GTT TGC AGT CAG AT | 291 bp | 60 | 45 |
| Reverse: GGT ACC CTT GCC CTA GTA AT | ||||
| TTR2 | Forward: TGG TGG AAA TGG ATC TGT CTG | 333 bp | 60 | 45 |
| Reverse: TGG AAG GGA CAA TAA GGG AAT |
EGFR, epidermal growth factor receptor; PCR, polymerase chain reaction; Ta, annealing temperature; TTR, transthyretin.
PCR was performed in a 50 μl final volume using standard conditions. Every reaction included 250 ng of DNA, 15 pmoles of each primer, 50mM KCl, 1.5mM MgCl2, 10mM Tris/HCl (pH 9 at room temperature), 200 μM of each dNTP, and 1 U of Taq DNA polymerase (Amersham Biosciences). The following amplification programme was used for all amplifications: denaturation at 95°C for three minutes; five cycles of 95°C for one minute, 60°C for one minute, and 72°C for one minute; then 45 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. The lengths of the amplicons were 90 bp for EGFR (GeneBank X17054), 204 bp for p16 (GeneBank NM_000077), 291 bp for TTR1 (GeneBank M11844), and 333 bp for TTR2 (GeneBank M11844).
Aliquots (10 μl) of the amplification products were run on 8% polyacrylamide gels. Gels were stained with ethidium bromide (5 μg/ml) and recorded using a VersaDoc imaging system (Bio-Rad Laboratories, Hercules, California, USA).
RT-PCR for RNA analysis
DNase I digestion was performed to eliminate genomic DNA from the RNA solution. The RNA (2 μg) was resuspended in 20 μl final volume containing 1× DNase I buffer, 80 U of RNase inhibitor (Ambion, Austin, Texas, USA), and 10 U of I RNase free DNase (Amersham Biosciences). The reaction was left to proceed at 37°C for 20 minutes. To purify RNA from DNase I an extraction with phenol/H2O/chloroform was performed. The final concentration of RNA in the solution was made by precipitation with isopropanol using 5 μl of glycogen (1 mg/ml) as precipitation carrier because of its low concentration.10,11
Reverse transcription was performed, as described previously,13 using 500 ng of total RNA, AMV reverse transcriptase (Promega; Madison, Wisconsin, USA), and sequence specific antisense primer. Table 2 shows the primers and PCR conditions used for RNA analysis.
Table 2.
Sequence of primers and PCR conditions for RNA analysis
| Gene | Primer sequence 5′–3′ | Length of product | Ta | Cycles |
| β Actin | Forward: ATC ACT GCC CTG GCA CCC A | 77 bp | 60 | 30 |
| Reverse: CCG ATC CAC ACG GAG TAC TTG | ||||
| β Actin | Forward: 5TTG CCG ACA GGA TGC AGA A | 100 bp | 60 | 30 |
| Reverse: the same primer used for β-Act 77 bases | ||||
| β Actin | Forward: TGG AGA AGA GCT ACG AGC TG | 120 bp | 58 | 40 |
| Reverse : GAA GGT AGT TTC GTG CAT CG | ||||
| β Actin | Forward: CTG GAC TTC GAG CAA GAG AT | 170 bp | 58 | 40 |
| Reverse: the same primer used for β-Act 120 bases | ||||
| β Actin | Forward: GAG AAG CTG TCC TAC GTC G-3′ | 200 bp | 59 | 45 |
| Reverse: the same primer used for β-Act 120 bases | ||||
| EGFR | Forward: GGC TCT GGA GGA AAA GAA AG | 65 bp | 55 | 40 |
| Reverse: TCA AAA GTG CCC AAC TGC TG | ||||
| EGFR | Forward: AAC ATC CTC TGG AGG CTG A | 95 bp | 55 | 40 |
| Reverse: TCA AAA GTG CCC AAC TGC TG | ||||
| GAPDH | Forward: CCC TCA ACG ACC ACT TTG TCA A | 75 bp | 58 | 30 |
| Reverse: GGT CCA CCA CCC TGT TGC T | ||||
| GAPDH | Forward: TCC ACC TTT GAC GCT GGG | 103 bp | 58 | 30 |
| Reverse: GGT CCA CCA CCC TGT TGC T |
EGFR, epidermal growth factor receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RT-PCR, reverse transcription polymerase chain reaction; Ta, annealing temperature.
For β actin (GeneBank M10277), we analysed five systems characterised by increasing length of the RT-PCR products (77, 100, 120, 170, and 200 bp) and for EGFR (GeneBank AF124513) two systems were evaluated (65 and 95 bp). We also analysed two systems (75 and 103 bp) for the human glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH; GeneBank NM_002046). Aliquots (10 μl) of each PCR product were visualised on 8% polyacrylamide gels. Gels were stained with ethidium bromide (5 μg/ml) and recorded using a VersaDoc imaging system (Bio-Rad Laboratories).
RESULTS
Twenty Bouin’s fixed, paraffin wax embedded breast cancer tissues were submitted to DNA and RNA extraction. Nucleic acids were extracted from 6 μm sections of paraffin wax embedded blocks. Ten sections were cut from every sample. Means of 257 ng of DNA and 51 ng of RNA were obtained for each 6 μm section. Every PCR and RT-PCR analysis was repeated twice to confirm the reproducibility of the results.
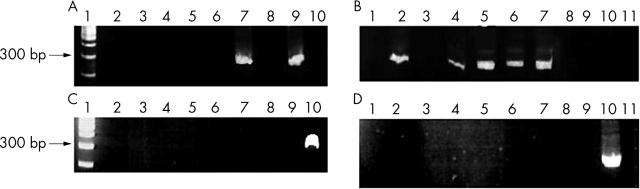
DNA samples were amplified with and without the restoration step (table 3). EGFR (100 bp) DNA was amplified in all the treated samples and in 18 of the 20 untreated samples (table 3). For PCR fragments of 204 bp (p16), the number of positive amplifications decreased only in the untreated samples (table 3; 15 of 20). For even longer amplification stretches of around 300 bp (TTR1, 291 bp; TTR2, 333 bp), no amplification was seen in untreated samples. In treated samples, the number of positive results decreased with the length of amplicons. For TTR1 (table 3; fig 1), seven treated samples were positively amplified and for TTR2 only two treated samples were amplified (fig 1).
Table 3.
DNA amplification results
| Gene (amplicon size) | Untreated DNA* | Treated DNA† |
| EGFR (100 bp) | 18/20 | 20/20 |
| p16 (204 bp) | 15/20 | 20/20 |
| TTR1 (291 bp) | 0/20 | 7/20 |
| TTR2 (333 bp) | 0/20 | 2/20 |
*DNA not subjected to the restoration step; † DNA subjected to the restoration step.
EGFR, epidermal growth factor receptor; TTR, transthyretin gene.
Figure 1.
PCR products for TTR1 and TTR2 analysis in Bouin’s solution fixed, paraffin wax embedded tissues. (A) TTR1 DNA (291 bp) with the restoration step. Lane 1, molecular size marker; lanes 2–10, DNA from breast cancer cases. (B) TTR1 DNA (291 bp) with the restoration step. Lanes 1–11, DNA from breast cancer cases. (C) TTR2 gene DNA (333 bp) with the restoration step. Lane 1, molecular size marker; lanes 2–10, DNA from breast cancer cases. (D) TTR gene DNA (333 bp) with the restoration step. Lanes 1–11, DNA from breast cancer cases. PCR, polymerase chain reaction; TTR, transthyretin gene.
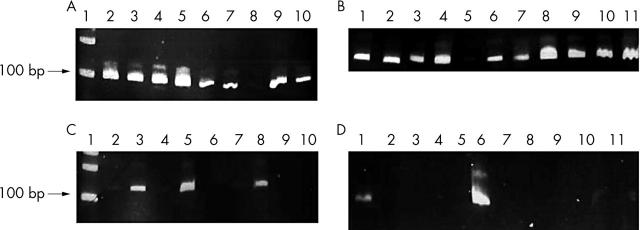
For RNA analysis (table 4), all amplicons that amplified successfully were no longer than 65–75 bp (EGFR, 65 bp; GAPDH, 75 bp; β actin, 77 bp). For longer fragments, the positivity rate decreased. For EGFR (95 bp), β actin (100 bp), and GAPDH (103 bp), 17 of 20, 18 of 20, and 16 of 20 amplicons were amplified, respectively. Increasing the length of the RT-PCR products decreased the number of positive results to seven of 20 samples. No results were obtained for the β actin RT-PCR products 170 and 200 bp long.
Table 4.
RNA amplification results
| Gene | RT-PCR results |
| β Actin (77 bp) | 20/20 |
| β Actin (100 bp) | 18/20 |
| β Actin (120 bp) | 7/20 |
| β Actin (170 bp) | 0/20 |
| β Actin (200 bp) | 0/20 |
| EGFR (65 bp) | 20/20 |
| EGFR (95 bp) | 17/20 |
| GAPDH (75 bp) | 20/20 |
| GAPDH (103 bp) | 16/20 |
EGFR, epidermal growth factor receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RT-PCR, reverse transcription polymerase chain reaction.
DISCUSSION
Nucleic acid degradation depends on environmental factors such as enzymes and temperature, in addition to chemical factors related to fixative solutions. Different fixatives have a different capacity to degrade DNA and RNA.9 It has already been shown that DNA from formalin fixed, paraffin wax embedded tissues can be reconstructed to longer fragments,8,14 but many doubts have been expressed about the possibility of DNA analysis in Bouin’s fixed tissues. Bouin’s fixative solution has been particularly recommended for testicular and lymphoma biopsies and for historical reasons has also been used in French and Canadian laboratories. Several authors have reported the difficulties related to molecular analysis in Bouin’s fixed tissues.6,15 Here, we show that DNA from Bouin’s fixed tissues can be reconstructed and analysed, but to a shorter length than formalin fixed tissues.8
“We recommend analysing 100–200 bp fragments of DNA from Bouin’s fixed tissues to guarantee amplification, and preferably to use a DNA reconstruction step”
We amplified DNA samples derived from Bouin’s fixed tissue sections, with and without a restoration step (table 3). Every PCR and RT-PCR analysis was repeated twice to confirm the reproducibility of the results, which were always consistent. As reported, EGFR DNA (100 bp) was amplified in all treated but in fewer untreated samples. PCR analysis in DNA from Bouin’s fixed tissue sections was successful in all samples that had been submitted to reconstruction for fragments up to 100–200 bp. For longer fragments, amplification was successful in only seven of 20 samples using amplicon sizes of 290 bp and only two of 20 samples of 330 bp, but only after DNA reconstruction. The inclusion of picric and acetic acid in Bouin’s fixative enhances DNA degradation in Bouin’s fixed tissues compared with formalin fixed tissues, in which over 300 bp can be amplified.5 Therefore, we recommend analysing 100–200 bp fragments of DNA from Bouin’s fixed tissues to guarantee amplification, and preferably to use a DNA reconstruction step.8
As already reported,10 it is possible to extract and analyse RNA from formalin fixed, paraffin wax embedded tissues. In routinely treated formalin fixed material, fragments of RNA between 100 and 200 bp can be amplified. RNA obtained from Bouin’s fixed, paraffin wax embedded tissues showed a higher degree of degradation and only amplicons less than 100 bp could be successfully amplified (table 4). For fragments around 100 bp long, RNA amplification was successful in a smaller number of cases (β actin, 18 of 20 cases; EGFR, 17 of 20 cases; GAPDH, 16 of 20 cases). For longer RT-PCR fragments of about 120 bp only seven of 20 samples were positive. All cases were negative when fragments of 170–200 bp were analysed, showing the limited length of amplicons that can be used in RT-PCR analysis of Bouin’s fixed tissues.
These findings show that Bouin’s fixative damages both DNA and RNA; however, it is possible to perform molecular analyses in these types of tissue by reducing the amplification fragment to less then 100 bp for RNA and less than 200 bp for DNA.
Take home messages.
Bouin’s fixative damages both DNA and RNA
Molecular analyses can be carried out in Bouin’s fixed tissues if the amplification fragment is reduced to less then 100 bp for RNA and less than 200 bp for DNA
Figure 2.
RT-PCR products in Bouin’s solution fixed, paraffin wax embedded tissues. (A) mRNA analysis for the β actin gene (100 bp). Lane 1, molecular size marker; lanes 2–10, RNA obtained from breast cancer cases. (B) mRNA analysis for β actin gene (100 bp). Lanes 1–11, RNA obtained from breast cancer cases. (C) mRNA analysis for β actin gene (120 bp). Lane 1, molecular size marker; lanes 2–10, RNA obtained breast cancer cases. (D) mRNA analysis for β actin gene (120 bp). Lanes 1–11, RNA obtained from breast cancer cases. RT-PCR, reverse transcription polymerase chain reaction.
Acknowledgments
We thank Ms S Vincent and Dr S Miertusova for the English revision of the manuscript.
Abbreviations
EGFR, epidermal growth factor receptor
GAPDH, human glyceraldehyde-3-phosphate dehydrogenase
PCR, polymerase chain reaction
RT, reverse transcription
TTR, transthyretin gene
The first two authors contributed equally to this study
REFERENCES
- 1.Luna LG. Manual of histologic staining methods—AFIP. 3rd ed. New York: Mc Graw-Hill, 1968.
- 2.Wollina U, Schreiber G, Zollmann C, et al. Lectin-binding sites in normal human testis. Andrologia 1989;21:127–30. [PubMed] [Google Scholar]
- 3.Konishi N, Ward JM, Reynolds CW, et al. Thymic T-cell lymphoma with the CD8+ (OX-8), CD4+ (W3/25) phenotype, induced in F344/NCr rats by nitroso-2-hydroxypropylurea. Thymus 1988;12:225–37. [PubMed] [Google Scholar]
- 4.Greer CE, Peterson SL, Kiviat NB, et al. PCR amplification from paraffin-embedded tissues. Effects of fixative and fixation time. Am J Clin Pathol 1991;95:117–24. [DOI] [PubMed] [Google Scholar]
- 5.Gall K, Pavelic J, Jadro-Santel D, et al. DNA amplification by polymerase chain reaction from brain tissues embedded in paraffin. Int J Exp Pathol 1993;74:333–7. [PMC free article] [PubMed] [Google Scholar]
- 6.Tyrrell L, Elias J, Longley J. Detection of specific mRNAs in routinely processed dermatopathology specimens. Am J Dermatopathol 1995;17:476–83. [DOI] [PubMed] [Google Scholar]
- 7.Biagini P, Monges G, Cantaloube JF, et al. Detection of gastrin mRNA in paraffin-embedded samples of normal antral mucosae using polymerase chain reaction technique. APMIS 1994;102:526–32. [DOI] [PubMed] [Google Scholar]
- 8.Bonin S, Petrera F, Niccolini B, et al. PCR analysis in archival postmortem tissues. Mol Pathol 2003;56:184–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Ezra J, Johnson DA, Rossi J, et al. Effect of fixation on the amplification of nucleic acids from paraffin-embedded material by the polymerase chain reaction. J Histochem Cytochem 1991;39:351–4. [DOI] [PubMed] [Google Scholar]
- 10.Stanta G, Schneider C. RNA extracted from paraffin-embedded human tissues is amenable to analysis by PCR amplification. Biotechniques 1991;11:304, 306, 308. [PubMed] [Google Scholar]
- 11.Stanta G, Bonin S, Perin R. RNA extraction from formalin-fixed and paraffin-embedded tissues. Methods Mol Biol 1998;86:23–6. [DOI] [PubMed] [Google Scholar]
- 12.Trevisan G, Stinco G, Nobile C, et al. Detection of Borrelia burgdorferi in skin biopsies from patients with morphea by polymerase chain reaction. J Eur Acad Dermatol Venereol 1996;6:15–19. [Google Scholar]
- 13.Bonin S, Pascolo L, Croce LS, et al. Gene expression of ABC proteins in hepatocellular carcinoma, perineoplastic tissue, and liver diseases. Mol Med 2002;8:318–25. [PMC free article] [PubMed] [Google Scholar]
- 14.Imyanitov EN, Grigoriev MY, Gorodinskaya VM, et al. Partial restoration of degraded DNA from archival paraffin-embedded tissues. Biotechniques 2001;31:1000, 1002. [DOI] [PubMed] [Google Scholar]
- 15.Tbakhi A, Totos G, Pettay JD, et al. The effect of fixation on detection of B-cell clonality by polymerase chain reaction. Mod Pathol 1999;12:272–8. [PubMed] [Google Scholar]