Abstract
Aim: To gain more insight into the genes involved in the aetiology and pathogenesis of anaplastic large cell lymphoma (ALCL).
Methods: Serial analysis of gene expression (SAGE) was undertaken on the CD4+ALK+ (anaplastic lymphoma kinase positive) ALCL derived cell line Karpas299 and as comparison on CD4+ T cells. Quantitative reverse transcription polymerase chain reaction (RT-PCR) and immunohistochemistry were performed on five ALCL derived cell lines and 32 tissue samples to confirm the SAGE data.
Results: High expression of Mcl-1 was seen in the Karpas299 cell line, whereas the two other antiapoptotic Bcl-2 family members, Bcl-2 and Bcl-XL, were not detected in the SAGE library. Quantitative RT-PCR confirmed the high expression of Mcl-1 mRNA and low expression of Bcl-2 and Bcl-XL in Karpas299 and in four other ALCL cell lines. To expand on these initial observations, primary tissue samples were analysed for Mcl-1, Bcl-XL, and Bcl-2 by immunohistochemistry. All 23 ALK+ and nine ALK− ALCL cases were positive for Mcl-1. Bcl-2 and Bcl-XL were expressed infrequently in ALK+ ALCL cases, but were present in a higher proportion of ALK− ALCL cases.
Conclusion: The consistent high expression of Mcl-1 in ALK+ and ALK− ALCL suggests that Mcl-1 is the main antiapoptotic protein in this disease. The high frequency of Mcl-1, Bcl-2, and Bcl-XL positive ALCL cases in the ALK− group compared with the ALK+ group indicates that ALK induced STAT3 activation is not the main regulatory pathway in ALCL.
Keywords: anaplastic large cell lymphoma, ALK, Bcl-2, Bcl-X L, Mcl-1
Anaplastic large cell lymphoma (ALCL) is characterised by the presence of a cohesive proliferation of large CD30 positive cells with abundant cytoplasm and pleomorphic, often horseshoe shaped nuclei.1 Most ALCL cases are of the T cell type, but null cell type ALCL also occurs.2,3 Two forms of ALCL can be distinguished based on the presence or absence of translocations involving the anaplastic lymphoma kinase (ALK) gene, such as the (2;5)(p23;q35) translocation (NPM/ALK).4,5 More recently, variant translocation partners have been described that also lead to overexpression of the ALK protein.1 ALK positive cases occur more frequently in children and young adults and have a relatively good prognosis with appropriate chemotherapy. ALK negative ALCL occurs in older individuals and has a poor prognosis.6,7
“Mcl-1 is a member of the Bcl-2 family of apoptosis regulating proteins (including Bcl-2, Bcl-XL, Bcl-XS, Bax, Bak, and Bad) and can inhibit apoptotic cell death”
Little is known about the biology of this malignant disease and only few studies have been published identifying genes specifically expressed in ALCL.8,9,10,11 To gain more insight into the genes that are involved in the aetiology and pathogenesis of ALCL, we applied serial analysis of gene expression (SAGE).12 This technique allows the construction of a comprehensive expression profile and results in a quantitative overview of the expression of the genes corresponding to the SAGE tags. Comparison of the CD4+ALK+ ALCL derived cell line Karpas299 with flow cytometry sorted peripheral blood CD4+ T cells revealed high expression of Mcl-1 in Karpas299. Mcl-1 is a member of the Bcl-2 family of apoptosis regulating proteins (including Bcl-2, Bcl-XL, Bcl-XS, Bax, Bak, and Bad) and can inhibit apoptotic cell death.13,14 Mcl-1 is also involved in the programming of differentiation,15 and promotes cell viability.16 In contrast to the apoptosis inhibiting potential of the full length Mcl-1 transcript, it has been suggested that differential splicing of the Mcl-1 gene yields a shorter protein with a Bcl-2 homology domain 3 that may promote cell death.17
To study Mcl-1 and other antiapoptotic Bcl-2 family members Bcl-2 and Bcl-XL we performed real time reverse transcription polymerase chain reaction (RT-PCR) and immunohistochemistry on ALK+ and ALK− ALCL cases.
MATERIALS AND METHODS
Cell lines and tissues
CD4+ T cells were isolated from the buffy coats of healthy donors using a fluorescence activated cell sorter (MoFlo Cytomation, Fort Collins, Colorado, USA) and were used directly for RNA isolation and SAGE analysis. The CD4+ALK+ ALCL derived cell line Karpas299 was obtained from ATCC (Rockville, Maryland, USA) and the NPM/ALK positive ALCL derived cell lines SU-DHL-1, SR786, SUP-M2, and DEL were obtained from DSMZ (Braunschweig, Germany). Based on the presence of the t(2;5) translocation, expression of CD4 and ALK, and lack of expression of B cell markers, we selected the Karpas299 cell line for the SAGE analysis. Frozen and paraffin wax embedded ALCL tissue specimens from 23 ALK+ and nine ALK− ALCL cases were obtained from the departments of pathology of the Groningen University Medical Centre, the Netherlands and the University of Malaysia. All protocols for obtaining and studying human tissues and cells were approved by the institution’s review board for human subject research.
SAGE
A detailed protocol for the SAGE procedure and a computer program (SAGE2000 version 4.12) for the analysis of gene specific tags were kindly provided by Dr KW Kinzler (John Hopkins Oncology Center, Baltimore, Maryland, USA).12 The SAGE procedure was performed as described previously.18 The SAGE libraries were compared and linked to the Unigene library to identify the corresponding genes.
Gene specific real time RT-PCR
Total RNA was isolated with Trizol (Life Technologies Inc, Gaithersburg, Maryland, USA) and first strand cDNA synthesis was performed using the protocol provided by the manufacturer (Life Technologies Inc). Assays-on-Demand™ gene expression products (Applied Biosystems, Foster City, California, USA) were used for Bcl-2 (Hs00236808_s1), Bcl-XL (Hs00236329_m1), and Mcl-1 (Hs00172031_m1). Real time PCR was performed in 1× Taqman® Universal PCR master mix (Applied Biosystems). β2 Microglobulin (β2m) was used as a positive control and for normalisation; β2m forward (5′-gaaaaagtggagcattcagacttg-3′), β2m reverse (5′-atgatgctgcttacatgtctcgat-3′), and probe (5′-agtcacatggttcacacggcaggc-3′) were dual labelled with 6-carboxyfluorescein (FAM) and 6-carboxytetramethylrhodamine (TAMRA). PCR was performed in triplicate in 1× qPCR master mix (Eurogentec, Liege, Belgium), using 900nM primers and 200nM probe. Reactions were performed on an ABI7900HT Sequence Detection System device (PE Applied Biosystems) using the standard program. Fluorescence was measured by means of sequence detection system software (SDS; version 2.0; Applied Biosystems). Mean cycle threshold values (Ct) and SDs were calculated for all genes. The amounts of the Bcl-2, Bcl-XL, and Mcl-1 targets were normalised relative to the amount of β2m target (ΔCt = ΔCt(gene) − ΔCt(β2m)) and the SD of ΔCt (SD(ΔCt)) was calculated (SD(ΔCt) = √((SDgene)2 + (SDβ2m)2). The relative amount of target gene was measured by determining ΔΔCt (ΔΔCt = ΔCtcalibrator − ΔCttestsample) and the factor difference was calculated (2−ΔΔCt). The range is given as 2−ΔΔCt+SDΔCt and 2−ΔΔCt−SDΔCt.
Immunostaining
ALK, Bcl-2, Bcl-XL, and Mcl-1 positivity was assessed by immunohistochemistry using standard protocols and appropriate dilutions of monoclonal mouse antihuman ALK antibody, monoclonal mouse antihuman Bcl-2 antibody, polyclonal rabbit antihuman Mcl-1 antibody (Dako, Copenhagen, Denmark), and monoclonal rabbit antihuman Bcl-XL antibody (Zymed, San Francisco, California, USA). Peroxidase activity was visualised with diaminobenzidine and H2O2. For all analyses, negative controls (first incubation step without primary antibody) and positive control tissue sections were included. Differences in the number of positive cases between the ALK+ and ALK− group were tested using Fisher’s exact test.
RESULTS
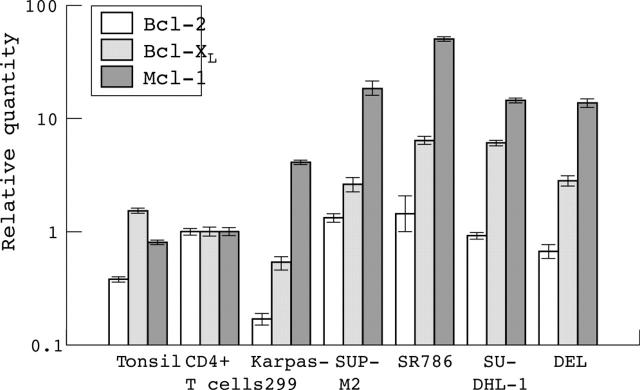
We constructed gene expression profiles of Karpas299 and CD4+ T cells using the SAGE technique. For Karpas299 we sequenced 10 678 tags representing 5090 different genes and for the CD4+ T cells we obtained 8425 tags representing 4467 different genes. These expression profiles were compared and linked to the Unigene library to identify the corresponding genes. The SAGE tag belonging to the Mcl-1 gene was detected at a frequency of 0.04% in Karpas299, and 0.01% in the CD4+ T cells. The tag for the shorter splice variant of Mcl-117 was not detected in the SAGE libraries. No SAGE tags were seen for the other antiapoptotic members of the Bcl-2 family (Bcl-2 and Bcl-XL) in Karpas299 or in CD4+ T cells. To confirm this differential expression, ALK+ ALCL cell lines Karpas299, SUP-M2, SR786, SU-DHL-1, and DEL were analysed using β2m, expressed at similar levels based on the SAGE libraries (0.29% in Karpas299 and 0.24% in CD4+ T cells), as a housekeeping gene. All five cell lines highly expressed Mcl-1 compared with tonsil and CD4+ T cells. Bcl-XL expression was moderate in the SUP-M2, SR786, SU-DHL-1, and DEL cell lines but was low in Karpas299 compared with CD4+ T cells and tonsil. Bcl-2 was slightly upregulated in the SUP-M2 and SR786 cell lines, but was reduced in the other ALCL cell lines compared with the CD4+ T cells (fig 1).
Figure 1.
Real time reverse transcription polymerase chain reaction results for Bcl-2, Bcl-XL, and Mcl-1 in tonsil, CD4+ T cells, and the Karpas299, SUP-M2, SR786, SU-DHL-1, and DEL cell lines. The bars on a logarithmic scale indicate the relative amount of mRNA. It is clear that Mcl-1 is the main antiapoptotic protein expressed in anaplastic large cell lymphoma derived cell lines.
Immunocytochemical staining for Mcl-1 revealed strong positivity in the Karpas299 cell line (fig 2A) and positive cytoplasmic staining in most of the tumour cells in all ALK+ and ALK− ALCL cases. Staining for Bcl-2 was negative in all ALK+ cases, whereas four of the nine ALK− cases were positive. Bcl-XL was positive in eight of nine ALK− cases, but only in three of 18 ALK+ cases (table 1; fig 2). The ALK− group included significantly more Bcl-2 (p = 0.004) and Bcl-XL (p = 0.000) positive cases than the ALK+ group.
Figure 2.
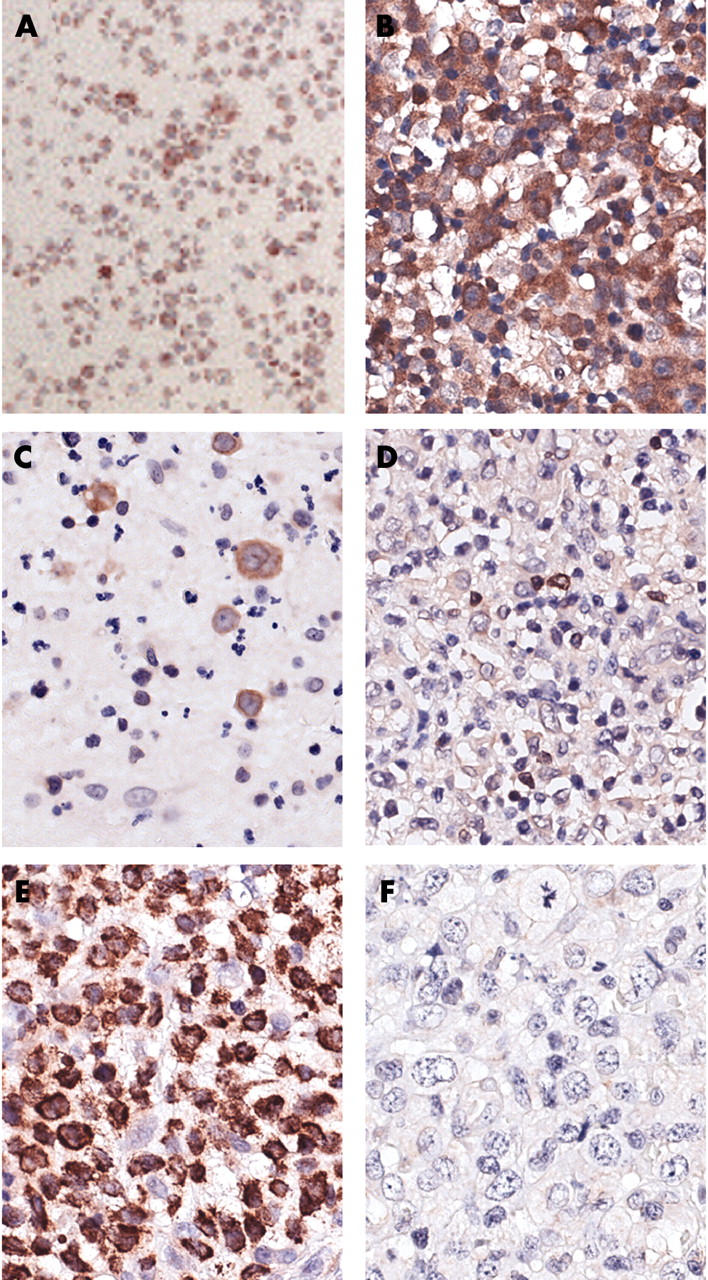
Immunohistochemistry for Mcl-1 in anaplastic large cell lymphoma (ALCL). (A) Mcl-1 staining in the Karpas299 cell line. (B) Mcl-1 staining in an anaplastic lymphoma kinase negative (ALK−) ALCL (case 31). (C) ALK− ALCL (case 25) with tumour cells positive for Bcl-2. (D) ALK− ALCL (case 23) with tumour cells negative for Bcl-2. (E) ALK− ALCL (case 28) with tumour cells positive for Bcl-XL. (F) ALK+ ALCL (case 8) with tumour cells negative for Bcl-XL. Original magnification, ×800.
Table 1.
Overview of the immunohistochemistry results of ALK+ and ALK− ALCL cases
| ALCL case | ALK | Mcl-1 | Bcl-2 | Bcl-XL |
| 1 | + | + | – | – |
| 2 | + | + | – | NR |
| 3 | + | + | – | NR |
| 4 | + | + | – | NR |
| 5 | + | + | – | NR |
| 6 | + | + | – | NR |
| 7 | + | + | – | – |
| 8 | + | + | – | – |
| 9 | + | + | – | + |
| 10 | + | + | – | – |
| 11 | + | + | – | – |
| 12 | + | + | – | + |
| 13 | + | + | – | – |
| 14 | + | + | – | – |
| 15 | + | + | – | – |
| 16 | + | + | – | – |
| 17 | + | + | – | – |
| 18 | + | + | – | – |
| 19 | + | + | – | – |
| 20 | + | + | – | – |
| 21 | + | + | – | – |
| 22 | + | + | – | + |
| 23 | + | + | – | – |
| 24 | – | + | – | + (few cells) |
| 25 | – | + | + | + |
| 26 | – | + | + | + |
| 27 | – | + | – | + (part of cells) |
| 28 | – | + | + | ++ |
| 29 | – | + | + | + |
| 30 | – | + | – | – |
| 31 | – | + | – | + (few cells) |
| 32 | – | + | – | + (part of cells) |
–, No staining in the tumour cells; +, moderate staining in most of the tumour cells; ++, strong staining in most tumour cells; NR, no result; few cells, staining in <10% of the tumour cells; part of cells, staining in 10–30% of the tumour cells.
ALCL, anaplastic large cell lymphoma; ALK, anaplastic lymphoma kinase.
DISCUSSION
Using SAGE analysis, the antiapoptotic member of the Bcl-2 family Mcl-1 was shown to be highly expressed in the ALCL derived cell line Karpas299 compared with CD4+ T cells. In contrast, tags corresponding to the other two members of the antiapoptotic Bcl-2 family, Bcl-XL and Bcl-2, were not identified in the SAGE libraries. Screening of the SAGE library for tags corresponding to proapoptotic genes revealed the presence only of a low frequency of tags corresponding to the Bak gene. The SAGE results were confirmed with quantitative RT-PCR, which indicated increased expression of the Mcl-1 gene in Karpas299 and in the four other ALCL derived cell lines. Moreover, quantitative RT-PCR for Bcl-XL and Bcl-2 clearly showed that expression was much lower than that of Mcl-1, consistent with the SAGE results.
Immunohistochemical analysis of 32 ALCL cases (23 ALK+ and nine ALK−) demonstrated the presence of the Mcl-1 protein in most tumour cells in all cases. No difference in Mcl-1 staining intensity or percentage of positive tumour cells was seen between ALK+ and ALK− cases. Mcl-1 protein expression was reported previously in 10 of 10 and 10 of 11 ALCLs.19,20 In a more recent study, Rasidakis et al detected Mcl-1 positivity in 16 of 26 ALK− ALCLs using a 10% cutoff.21 These data confirm the consistent high expression of Mcl-1 and strongly suggest that Mcl-1, rather than Bcl-2 or Bcl-XL, is the main antiapoptotic protein of the Bcl-2 family expressed in ALCL, and in particular in ALK+ ALCL. Analysis of the apoptotic rate in ALCL revealed low levels of apoptotic cells, ranging from 1.2% to 3.2%, in ALK+ and ALK− cases.21,22
A possible role for Mcl-1 in lymphomagenesis is supported by the finding of a variety of lymphomas in Mcl-1 transgenic mice.23 Moreover, high Mcl-1 expression has been reported in various other lymphoma subtypes, including angioimmunoblastic T cell lymphoma, myeloma cell lines, cutaneous T cell lymphoma, diffuse large B cell lymphoma, and mantle cell lymphoma.19,20,24,25,26,27 Treatment with the cyclin dependent kinase inhibitor flavopiridol, which results in a strong downregulation of Mcl-1 expression, has been shown to be effective in multiple myeloma, leukaemia cell lines, and in chronic lymphocytic leukaemia samples,28,29,30 indicating the importance of Mcl-1 as an antiapoptotic protein.
“Our findings argue against a role for ALK induced activation of STAT3 and suggest involvement of other pathways leading to the induction of Bcl-XL and Bcl-2 expression”
The inhibition of JAK3 in two ALK+ ALCL derived cell lines resulted in downregulation of activated STAT3, decreased levels of Bcl-2 and Bcl-XL, and no changes in Mcl-1 values.31 This suggests that other mechanisms are involved in the induction of Mcl-1 in ALCL cases. However, in a more recent study Amin et al demonstrated the downregulation of Bcl-2, Bcl-XL, and Mcl-1 upon transfection of a dominant negative STAT3 in two ALCL cell lines, supporting the STAT3 mediated induction of Mcl-1 in these cells.32 Similar results were obtained in macrophages and large granular lymphocyte leukaemia, which showed reduced Mcl-1 expression upon treatment with JAK inhibitors.33,34 Our data indicate the presence of the Mcl-1 protein in all ALCL cases, independent of expression of the ALK protein, which suggests that besides the ALK induced activation of STAT3, other pathways might also contribute to the induction of Mcl-1 expression in ALCL cases that lack ALK expression. This is supported by two studies that show the involvement of the phosphatidylinositol 3-kinase pathway in the upregulation of Mcl-1 expression.33,35
In addition to positive regulation via survival signals such as activated STAT3 and the phosphatidylinositol 3-kinase pathway, Mcl-1 can also be downregulated via E2F1.36 Treatment with flavopiridol results in stabilisation of E2F1, which acts as a transcriptional repressor of Mcl-1, and induces an effective downregulation of Mcl-1. Based on the balance between survival pathways and the effectiveness of Mcl-1 downregulation upon flavopiridol treatment, beneficial effects might be achieved by treatment of ALCL with flavopiridol.
Take home messages.
Mcl-1 was consistently highly expressed in ALK+ and ALK− anaplastic large cell lymphomas (ALCL), suggesting that Mcl-1 is the main antiapoptotic protein in this disease
The high frequency of Mcl-1, Bcl-2, and Bcl-XL positive ALCL cases in the ALK− group compared with the ALK+ group indicates that ALK induced STAT3 activation is not the main regulatory pathway in ALCL
Treatment with cyclin dependent kinase inhibitors such as flavopiridol may provide a novel tool for the treatment of both ALK+ and ALK− patients with ALCL
As mentioned above, several studies suggest a role for ALK induced activation of STAT3 in the induction of Bcl-2 and Bcl-XL expression in ALCL cases.31,37–39 Indeed, the presence of activated STAT3 was demonstrated in most ALK+ cases, but also in approximately half of the ALK− cases.38 These data indicate that STAT3 activation correlates with, but is not strictly dependent on, ALK expression in ALCL. The analysis of Bcl-2 and Bcl-XL in ALCL demonstrated the complete absence of Bcl-2 in ALK+ cases and the expression of Bcl-2 in four of nine ALK− cases, whereas Bcl-XL was expressed in only three of 18 ALK+ versus eight of nine ALK− cases. These data indicate that Bcl-2 and Bcl-XL are only infrequently expressed in ALK+ ALCL, but are present in a higher proportion of ALK− ALCLs. An inverse relation between ALK and Bcl-2/Bcl-XL expression was previously noted in ALCL.20,40–42 These findings argue against a role for ALK induced activation of STAT3 and suggest involvement of other pathways leading to the induction of Bcl-XL and Bcl-2 expression.
In summary, similar amounts of Mcl-1 protein are expressed in both ALK− and ALK+ ALCLs, whereas the expression of Bcl-2 and Bcl-XL is limited to ALK– cases. Because activated STAT3 is not detectable in a small proportion of ALK+ cases and in half of the ALK− cases, it is very likely that other regulatory pathways are involved in the expression of these antiapoptotic Bcl-2 family members. The consistent expression of Mcl-1 as demonstrated in our study indicates that treatment with agents such as flavopiridol may provide a novel tool for the treatment of both ALK+ and ALK− patients with ALCL.
Acknowledgments
This study was supported by the Foundation for Pediatric Oncology Research Groningen (SKOG, grant number 99-04).
Abbreviations
ALCL, anaplastic large cell lymphoma
ALK, anaplastic lymphoma kinase
β2m, β2 microglobulin
Ct, threshold cycle
RT-PCR, reverse transcription polymerase chain reaction
SAGE, serial analysis of gene expression
REFERENCES
- 1.Stein H, Foss HD, Dürkop H, et al. CD30+ anaplastic large cell lymphoma: a review of its histopathologic, genetic, and clinical features. Blood 2000;96:3681–95. [PubMed] [Google Scholar]
- 2.O’Connor NT, Stein H, Gatter KC, et al. Genotypic analysis of large cell lymphomas which express the Ki-1 antigen. Histopathology 1987;11:733–40. [DOI] [PubMed] [Google Scholar]
- 3.Ohshima K, Kikuchi M, Masuda Y, et al. Genotypic and immunophenotypic analysis of anaplastic large cell lymphoma (Ki-1 lymphoma). Pathol Res Pract 1990;186:582–8. [DOI] [PubMed] [Google Scholar]
- 4.Rimokh R, Magaud JP, Berger F, et al. A translocation involving a specific breakpoint (q35) on chromosome 5 is characteristic of anaplastic large cell lymphoma (Ki-1 lymphoma). Br J Haematol 1989;71:31–6. [DOI] [PubMed] [Google Scholar]
- 5.Pittaluga S, Wlodarska I, Pulford K, et al. The monoclonal antibody ALK1 identifies a distinct morphological subtype of anaplastic large cell lymphoma associated with 2p23/ALK rearrangements. Am J Pathol 1997;151:343–51. [PMC free article] [PubMed] [Google Scholar]
- 6.Skinnider BF, Connors JM, Sutcliffe SB, et al. Anaplastic large cell lymphoma: a clinicopathologic analysis. Hematol Oncol 1999;17:137–48. [DOI] [PubMed] [Google Scholar]
- 7.Sherman CG, Zielenska M, Lorenzana AN, et al. Morphological and phenotypic features in pediatric large cell lymphoma and their correlation with ALK expression and the t(2;5)(p23;q35) translocation. Pediatr Dev Pathol 2001;4:129–37. [DOI] [PubMed] [Google Scholar]
- 8.Wellmann A, Thieblemont C, Pittaluga S, et al. Detection of differentially expressed genes in lymphomas using cDNA arrays: identification of clusterin as a new diagnostic marker for anaplastic large-cell lymphomas. Blood 2000;96:398–404. [PubMed] [Google Scholar]
- 9.Gaiser T, Thorns C, Merz H, et al. Gene profiling in anaplastic large-cell lymphoma derived cell lines with cDNA expression arrays. J Hematother Stem Cell Res 2002;11:423–8. [DOI] [PubMed] [Google Scholar]
- 10.Thorns C, Gaiser T, Lange K, et al. cDNA arrays: gene expression profiles of Hodgkin’s disease and anaplastic large cell lymphoma cell lines. Pathol Int 2002;52:578–85. [DOI] [PubMed] [Google Scholar]
- 11.Villalva C, Trempat P, Greenland C, et al. Isolation of differentially expressed genes in NMP-ALK-positive anaplastic large cell lymphoma. Br J Haematol 2002;118:791–8. [DOI] [PubMed] [Google Scholar]
- 12.Velculescu VE, Zhang L, Vogelstein B, et al. Serial analysis of gene expression. Science 1995;270:484–7. [DOI] [PubMed] [Google Scholar]
- 13.Tsujimoto Y, Shimizu S. Bcl-2 family: life-or-death switch. FEBS Lett 2000;466:6–10. [DOI] [PubMed] [Google Scholar]
- 14.Adams JM, Cory S. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem Sci 2001;26:61–6. [DOI] [PubMed] [Google Scholar]
- 15.Craig RW. MCL1 provides a window on the role of the BCL2 family in cell proliferation, differentiation and tumorigenesis. Leukemia 2002;16:444–54. [DOI] [PubMed] [Google Scholar]
- 16.Zhou P, Qian L, Kozopas KM, et al. Mcl-1, a Bcl-2 family member, delays the death of hematopoietic cells under a variety of apoptosis-inducing conditions. Blood 1997;89:630–43. [PubMed] [Google Scholar]
- 17.Bingle CD, Craig RW, Swales BM, et al. Exon skipping in Mcl-1 results in a Bcl-2 homology domain 3 only gene product that promotes cell death. J Biol Chem 2000;275:22136–46. [DOI] [PubMed] [Google Scholar]
- 18.Van den Berg A, Visser L, Poppema S. High expression of the CC-chemokine TARC in Reed-Sternberg cells: a possible explanation for the characteristic T-cell infiltrate in Hodgkin’s lymphoma. Am J Pathol 1999;154:1685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlaifer D, Krajewski S, Galoin S, et al. Immunodetection of apoptosis-regulating proteins in lymphomas from patients with and without human immunodeficiency virus infection. Am J Pathol 1996;149:177–85. [PMC free article] [PubMed] [Google Scholar]
- 20.Rassidakis GZ, Jones D, Lai R, et al. BCL-2 family proteins in peripheral T-cell lymphomas: correlation with tumour apoptosis and proliferation. J Pathol 2003;200:240–8. [DOI] [PubMed] [Google Scholar]
- 21.Rassidakis GZ, Lai R, McDonnell TJ, et al. Overexpression of Mcl-1 in anaplastic large cell lymphoma cell lines and tumors. Am J Pathol 2002;160:2309–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drakos E, Rassidakis GZ, Lai R, et al. Caspase-3 activation in systemic anaplastic large-cell lymphoma. Mod Pathol 2004;17:109–16. [DOI] [PubMed] [Google Scholar]
- 23.Zhou P, Levy NB, Xie H, et al. Mcl-1 transgenic mice exhibit a high incidence of B cell lymphoma manifested as a spectrum of histological subtypes. Blood 2001;97:3902–9. [DOI] [PubMed] [Google Scholar]
- 24.Amin HM, McDonnell TJ, Medeiros LJ, et al. Characterization of 4 mantle cell lymphoma cell lines. Arch Pathol Lab Med 2003;127:424–31. [DOI] [PubMed] [Google Scholar]
- 25.Jourdan M, Veyrune JL, Vos JD, et al. A major role for Mcl-1 antiapoptotic protein in the IL-6-induced survival of human myeloma cells. Oncogene 2003;22:2950–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai R, Rassidakis GZ, Medeiros LJ, et al. Expression of STAT3 and its phosphorylated forms in mantle cell lymphoma cell lines and tumours. J Pathol 2003;199:84–9. [DOI] [PubMed] [Google Scholar]
- 27.Zhang CL, Kamarashev J, Qin JZ, et al. Expression of apoptosis regulators in cutaneous T-cell lymphoma (CTCL) cells. J Pathol 2003;200:249–54. [DOI] [PubMed] [Google Scholar]
- 28.Kitada S, Zapata JM, Andreeff M, et al. Protein kinase inhibitors flavopiridol and 7-hydroxy-staurosporine down-regulate antiapoptosis proteins in B-cell chronic lymphocytic leukemia. Blood 2000;96:393–7. [PubMed] [Google Scholar]
- 29.Gojo I, Zhang B, Fenton RG. The cyclin-dependent kinase inhibitor flavopiridol induces apoptosis in multiple myeloma cells through transcriptional repression and down-regulation of Mcl-1. Clin Cancer Res 2002;8:3527–38. [PubMed] [Google Scholar]
- 30.Rosato RR, Almenara JA, Cartee L, et al. The cyclin-dependent kinase inhibitor flavopiridol disrupts sodium butyrate-induced p21WAF1/CIP1 expression and maturation while reciprocally potentiating apoptosis in human leukemia cells. Mol Cancer Ther 2002;1:253–66. [PubMed] [Google Scholar]
- 31.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res 2002;8:945–54. [PubMed] [Google Scholar]
- 32.Amin HM, McDonnell TJ, Ma Y, et al. Selective inhibition of STAT3 induces apoptosis and G1 cell cycle arrest in ALK-positive anaplastic large cell lymphoma. Oncogene 2004;23:5426–34. [DOI] [PubMed] [Google Scholar]
- 33.Epling-Burnette PK, Liu JH, Catlett-Falcone R, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest 2001;107:351–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, Ma Y, Cole SM, et al. Serine phosphorylation of STAT3 is essential for Mcl-1 expression and macrophage survival. Blood 2003;102:344–52. [DOI] [PubMed] [Google Scholar]
- 35.Wei LH, Kuo ML, Chen CA, et al. The anti-apoptotic role of interleukin-6 in human cervical cancer is mediated by up-regulation of Mcl-1 through a PI 3-K/Akt pathway. Oncogene 2001;20:5799–809. [DOI] [PubMed] [Google Scholar]
- 36.Ma Y, Cress WD, Haura EB. Flavopiridol-induced apoptosis is mediated through up-regulation of E2F1 and repression of Mcl-1. Mol Cancer Ther 2003;2:73–81. [PubMed] [Google Scholar]
- 37.Zamo A, Chiarle R, Piva R, et al. Anaplastic lymphoma kinase (ALK) activates STAT3 and protects hematopoietic cells from cell death. Oncogene 2002;21:1038–47. [DOI] [PubMed] [Google Scholar]
- 38.Khoury JD, Medeiros LJ, Rassidakis GZ, et al. Differential expression and clinical significance of tyrosine-phosphorylated STAT3 in ALK+ and ALK− anaplastic large cell lymphoma. Clin Cancer Res 2003;9:3692–9. [PubMed] [Google Scholar]
- 39.Amin HM, Medeiros LJ, Ma Y, et al. Inhibition of Jak3 induces apoptosis and decreases anaplastic lymphoma kinase activity in anaplastic large cell lymphoma. Oncogene 2003;22:5399–407. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura S, Shiota M, Nakagawa A, et al. Anaplastic large cell lymphoma: a distinct molecular pathologic entity: a reappraisal with special reference to p80 (NPM/ALK) expression. Am J Surg Pathol 1997;21:1420–32. [DOI] [PubMed] [Google Scholar]
- 41.Rassidakis GZ, Sarris AH, Herling M, et al. Differential expression of Bcl-2 family proteins in ALK-positive and ALK-negative anaplastic large cell lymphoma of T/null-cell lineage. Am J Pathol 2001;159:527–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villalva C, Bougrine F, Delsol G, et al. Bcl-2 expression in anaplastic large cell lymphoma. Am J Pathol 2001;158:1889–90. [DOI] [PMC free article] [PubMed] [Google Scholar]