Abstract
Background: Fluorescence in situ hybridisation (FISH) is useful for detecting specific chromosomal abnormalities in various tumours. In lymphomas, diagnosis is frequently made using paraffin wax embedded tissue. However, FISH performed under these conditions presents potential technical problems and difficulties in interpretation.
Aims: To show that FISH using tissue imprints and cytopreps or alternatively, bone marrow (BM) smears, constitutes an easy and rapid strategy to overcome these constraints.
Methods: The study comprised 46 patients with lymphoma. Sixty nine tissue imprints, cytopreps, or BM smears were analysed by FISH. Dual colour, dual fusion FISH probes were used to detect the t(8;14), t(11;14), and t(14;18) translocations, whereas a dual colour breakapart FISH probe was used to detect chromosomal translocations involving the BCL6 gene.
Results: Tissue imprints and cytopreps were successfully hybridised in all 52 cases, whereas hybridisation was successful in 16 of 17 archival BM smears. All patients could be analysed to identify either the presence or absence of chromosomal translocations.
Conclusions: The use of tissue imprints, cytopreps, or BM smears to identify chromosomal abnormalities by FISH is a rapid and useful ancillary approach for diagnostic purposes. Therefore, it could be used on a routine basis whenever fresh samples are available.
Keywords: lymphoma, tissue imprints, chromosomal abnormalities, fluorescence in situ hybridisation
Several distinct genetic abnormalities have been specifically associated with lymphoproliferative and myeloproliferative disorders.1 In the case of lymphomas, diagnosis is mainly established on morphological and immunohistochemical grounds using samples routinely processed in paraffin wax. However, in at least some cases the identification of specific genetic markers is crucial—for example, t(11;14)(q13;q32) in mantle cell lymphoma,2 t(14;18)(q32;q21) in follicular lymphoma,3 t(3;var)(q27;var) in follicular or diffuse large B cell lymphoma,1,3 and t(8;14)(q24;q32) in Burkitt lymphoma.4 Fluorescence in situ hybridisation (FISH) is a suitable method for detecting genetic abnormalities.5 Although FISH can be performed on dewaxed tissue sections, this technique remains potentially challenging. The scoring of individual nuclei is difficult because of cellular overlap and nuclear truncation. Moreover, fixation and embedding procedures may produce artefacts in the tissue, thereby interfering with DNA hybridisation.6 To overcome these disadvantages, we hereby propose to perform FISH as a routine test on tissue imprints or cytopreps and, alternatively, on BM smears. Under these circumstances, FISH can be rapidly and easily performed, overcoming the disadvantages of paraffin wax embedded samples.
“Several distinct genetic abnormalities have been specifically associated with lymphoproliferative and myeloproliferative disorders”
METHODS
Material
Because our study was primarily designed to explore the applicability of the FISH technique on cytological preparations, we decided to test the above translocations in a series of unselected consecutive B cell lymphomas diagnosed in our institution from January 2002 to January 2004.
We analysed 69 samples comprising fresh tissue imprints, cytopreps, and bone marrow (BM) smears obtained from 46 patients with lymphoma (table 1).
Table 1.
Clinical data, probes used, and FISH results
| Patient | Sample | Clinical status* | FISH analysis† | Final Dx | ||
| Probe | Pattern | Result | ||||
| 1 | LN | Dx | t(11;14) | 1R1G2F (78%) | + | MCL |
| 2 | Intestine | Rel (+68) | t(11;14) | 1R1G6F (100%) | +‡ | MCL |
| 3 | Soft tissue | Dx | t(11;14) | 1R1G2F (100%) | + | MCL |
| LN | Rel (+10) | t(11;14) | 1R1G2F (90.4%) | + | ||
| 4 | LN | Dx | t(11;14) | 1R1G2F (46.4%) | + | MCL |
| 5 | Soft tissue | Dx | t(11;14) | 1R1G2F (75%) | + | MCL |
| 6 | LN | Dx | t(11;14) | 2R2G | − | MCL |
| 7 | BM | Dx | t(14;18) | 2R3G (6.6%) | −‡ | |
| BM | Dx | t(11;14) | 1R1G2F (6%) | + | MCL | |
| LN | Rel (+70) | t(14;18) | 2R4G (63%) | −‡ | ||
| LN | Rel (+70) | t(11;14) | 1R2G2F (100%) | +‡ | ||
| 8 | BM | Dx | t(11;14) | 1R0G3F (40%) | +‡ | MCL |
| 9 | BM | Dx | t(11;14) | 3R3G3F (38%) | +‡ | MCL |
| 10 | BM | Dx | t(11;14) | NE | ||
| BM | Rel (+63) | t(11;14) | 1R1G2F (72%), 2R2G3F (15%) | +‡ | MCL | |
| 11 | BM | Dx | t(11;14) | 1R1G2F (62%), 2R1G2F (22%) | +‡ | MCL |
| 12 | BM | Dx | t(11;14) | 1R1G2F (39%) | + | MCL |
| 13 | LN | Dx | t(11;14) | 2R2G | − | |
| LN | Dx | t(3;var) | 1R1G1F (17.6%) | + | FL | |
| 14 | LN | Rel (+178) | t(14;18) | 1R1G2F (53%) | + | FL |
| 15 | LN | Dx | t(14;18) | 1R1G2F (15%), 1R1G3F (12.4%), 2R2G3F (15.6%) | +‡ | FL |
| 16 | Salivary | Rel (+13) | t(14;18) | 2R2G | − | FL |
| Salivary | Rel (+13) | t(3;var) | 0R0G2F | − | ||
| 17 | LN | Dx | t(14;18) | 1R1G2F (44%) | + | FL |
| 18 | LN | Rel (+39) | t(14;18) | 1R1G2F (70%) | + | FL |
| 19 | LN | Dx | t(14;18) | 1R1G2F (30%), 1R1G1F (23%) | +‡ | FL |
| 20 | LN | Rel (+116) | t(14;18) | 1R1G2F (10.5%) | + | FL |
| 21 | LN | Dx | t(14;18) | 1R1G2F (29%), 1R1G1F (15%), 2R2G2F (10%) | +‡ | FL |
| 22 | Soft tissue | Dx | t(14;18) | 1R1G2F (71%), 0R0V2F (11%) | +‡ | FL |
| 23 | LN | Rel (+91) | t(14;18) | 1R1G2F (16%) | + | FL |
| 24 | LN | Dx | t(14;18) | 1R1G2F (13%) | + | FL |
| 25 | LN | Dx | t(14;18) | 1R1G2F (58%), 1R1G1F (23%) | +‡ | FL |
| 26 | LN | Dx | t(14;18) | 1R1G2F (37%) | + | FL |
| 27 | LN | Dx | t(11;14) | 2R3G (62%) | −‡ | |
| LN | Dx | t(14;18) | 1R1G2F (59.4%) | + | FL | |
| 28 | LN | Dx | t(11;14) | 2R3G (28%) | −‡ | |
| LN | Dx | t(14;18) | 1R1G2F (31%) | + | FL | |
| 29 | LN | Dx | t(14;18) | 2R2G2F (60%), 1R1G3F (11%) | +‡ | FL |
| 30 | Skin | Rel (+90) | t(14;18) | 1R1G2F (17%) | + | FL |
| 31 | LN | Dx | t(14;18) | 2R2G | − | |
| LN | Dx | t(3;var) | 1R1G1F (12.4%) | + | FL | |
| 32 | BM | Dx | t(14;18) | 1R1G2F (14%) | + | FL |
| 33 | BM | Dx | t(14;18) | 1R1G2F (75.6%) | + | FL |
| 34 | BM | Dx | t(14;18) | 2R2G | − | FL |
| BM | F-up (+7) | t(3;var) | 0R0G2F | − | ||
| 35 | LN | Dx | t(8;14) | 2A2R2G | − | DLBCL |
| LN | Dx | t(3;var) | 0R0G2F | − | ||
| BM | Dx | t(8;14) | 2A2R2G | − | ||
| BM | Dx | t(3;var) | 0R0G2F | − | ||
| 36 | Testicle | Rel (+41) | t(14;18) | 4R2G (26%) | −‡ | |
| Testicle | Rel (+41) | t(8;14) | 2A2R2G | − | ||
| Testicle | Rel (+41) | t(3;var) | 1R1G1F (26%) | + | DLBCL | |
| 37 | CNS | Dx | t(14;18) | 2R2G | − | |
| CNS | Dx | t(8;14) | 2A2R2G | − | ||
| CNS | Dx | t(3;var) | 1R1G2F (55%) | +‡ | DLBCL | |
| 38 | LN | Dx | t(14;18) | 2R2G | − | DLBCL |
| LN | Dx | t(11;14) | 2R2G | − | ||
| LN | Dx | t(3;var) | 0R0G2F | − | ||
| 39 | Ascites | Dx | t(14;18) | 4R4G (73%), 8R8G(8%) | −‡ | DLBCL |
| Ascites | Dx | t(8;14) | 4A4R4G (82%), 8A8R8G (3%) | −‡ | ||
| Ascites | Dx | t(3;var) | 0R0G2F | − | ||
| 40 | Ascites | Dx | t(14;18) | 3R3G (51%), 3R4G (11%) | −‡ | |
| Pleural fluid | Dx | t(14;18) | 1R1G1F (25%), 3R3G (13%), 2R3G (19%) | +‡ | DLBCL | |
| 41 | LN | Dx | t(8;14) | 2A2R2G | − | DLBCL (HIV) |
| 42 | Soft tissue | Dx | t(14;18) | 2R2G | − | MZL |
| 43 | BM | Rel (+22) | t(11;14) | 2R2G | − | MZL |
| 44 | BM | Dx | t(11;14) | 2R2G | − | B-CLL |
| 45 | Ascites | Rel (+85) | t(11;14) | 2R2G | − | B-CLL |
| 46 | BM | Dx | t(11;14) | 2R2G | − | B-CLL |
*Figures in parenthesis show the time in months from diagnosis; †cf table 2; ‡atypical FISH pattern.
B-CLL, B chronic lymphocytic leukaemia; BM, bone marrow; CNS, central nervous system; DLBCL, diffuse large B cell lymphoma; Dx, diagnosis; FISH, fluorescence in situ hybridisation; FL, follicular lymphoma; F-up, follow up; HIV, human immunodeficiency virus; LN, lymph node; MCL, mantle cell lymphoma; MZL, marginal zone lymphoma; NE, not evaluated; Rel, relapse.
Slides were prepared conventionally, air dried for 24 hours, and stored at room temperature when FISH was to be performed within one week or at −20°C for longer storage periods. In addition, archival BM smears stored at room temperature for up to 12 years were also included in our study.
All diagnoses were previously proposed based upon conventional morphology, supplemented by immunocytochemistry and/or flow cytometry. To avoid false negative results by FISH, special care was taken to ensure the presence of tumour cells in each slide by rapid May-Grümwald Giemsa staining (Quick Panoptic QCA, Amposta, Spain).
Fluorescence in situ hybridisation
Fresh slides were aged at room temperature for at least 24 hours, fixed twice in fresh Carnoy (methanol/acetic acid; 3/1) for five minutes each, and dried at room temperature for a further 24 hours. FISH was performed according to the instructions of the probe supplier (Vysis Inc, Downers Grove, Illinois, USA), with minor modifications. Pretreatment of the slides included incubation in 2× saline sodium citrate (SSC) at 37°C for 30 minutes and in 0.005% pepsin/0.01N HCl at 37°C for one hour, in both cases followed by dehydration in a series of ethanol (70%, 85%, and 100% in distilled water) and air drying.
After adding the probe (table 2) and placing a coverslip, denaturation was performed using the metal block of a thermocycler (HyBrite, Vysis Inc) at 75°C for five minutes and gradually cooling to 37°C. Slides were hybridised overnight (16 hours) in a humid chamber at 37°C.
Table 2.
Characteristics of the probes used
| Probes (labelling) | Hybridisation loci | Anomaly detected | Typical FISH pattern | |||
| Normal | Abnormal | Type | Sensitivity | |||
| IGH (G), BCL2 (R) | 14q32, 18q21 | t(14;18)(q32;q21) | 2R2G | 1R1G2F | D-FISH | 1% |
| CCND1 (R), IGH (G) | 11q13, 14q32 | t(11;14)(q13;q32) | 2R2G | 1R1G2F | D-FISH | 1% |
| CEP8 (A), c-MYC (R), IGH (G) | 8p11.1–q11.1, 8q24, 14q32 | t(8;14)(q24;q32) | 2A2R2G | 2A1R1G2F | D-FISH | 1% |
| BCL6 (F) | 3q27 | t(3;var)(q27;var) | 0R0G2F | 1R1G1F | Breakapart | 4% |
Sensitivity for the detection of atypical patterns. D-FISH probes: 6.5% for 2R3G; 2.4% for 1R1G1F; 1% for 2R4G, 4R2G, 3R3G, 3R4G, 4R4G, 1R2G2F, 2R1G2F, 2R2G2F, and 0R0V2F; 0.6% for 8R8G, 1R0G3F, 1R1G3F, 3R3V3F, 2R2V3F, 1R1V6F, 4A4R4V, and 8A8R8V (maximum number of false positive cells in 500 cells scored was 23, 6, 1, and 0, respectively); “breakapart” probe: 0.6% for 1R1G2F (maximum number of false positive cells in 500 cells scored was 0).
A, aqua signal; D-FISH, dual colour dual signal fluorescence in situ hybridisation; F, red/green fusion signal; G, green signal; R, red signal.
After hybridisation, slides were washed in 2× SSC at 65°C for two minutes, followed by 0.1% NP-40/2× SSC for one minute at room temperature, and counterstained with 125 ng/ml DAPI (4,6-diamidine-2-phenyl indole; Vysis Inc).
The sensitivity of the FISH assay for the detection of the gene fusion or breakapart patterns was determined using the upper boundary of a one sided 95% confidence interval for observing the maximum number of false positive cells in 500 cells scored (one for dual colour, dual signal FISH patterns, 12 for breakapart patterns) found in 10 tissue imprints from normal tissue (palatine tonsils) with each probe (table 2).
In 14 cases from which only a single slide was available, FISH for different rearrangements was performed by sequentially rehybridising the same slide with different probes, as described previously.7 A similar approach was used to rehybridise (with the same probe) 10 samples in which unsatisfactory FISH signals were obtained in a first hybridisation attempt.
RESULTS
All tissue imprints and cytopreps were successfully hybridised with the protocol described above. All but one (patient number 10) BM smears, including those stored for long periods of time (up to 12 years) at room temperature, were also successfully hybridised. These results include those cases in which FISH with different probes was sequentially performed on the same slide (n = 14), and samples in which insufficient signal was obtained in a first FISH attempt that were successfully analysed after washing and rehybridising the sample with the same probe (n = 10).
Translocation t(11;14)
Twenty three samples from 20 patients (table 1) were studied for IGH–CCND1 rearrangements (table 2). Thirteen samples were positive and nine were negative. FISH was unsuccessful in one sample (BM smear stored for nine years; patient number 10). In this case, a positive confirmatory result was subsequently obtained from a more recent BM smear (stored for three years), corresponding to a relapse (table 1). Atypical FISH patterns were seen in six positive samples (possibly as a result of variant or complex translocations, simultaneously displaying two different clones in patients 10 and 11) and two negative cases (with a split IGH signal resulting from a t(14;18) translocation). FISH allowed the reclassification of one particular patient (patient number 7) from an initial diagnosis of follicular lymphoma to mantle cell lymphoma. Moreover, FISH performed on a BM smear from this patient at diagnosis disclosed a conventional pattern, whereas an atypical result was obtained on a fresh tissue imprint obtained at relapse (six years later) (fig 1). The remaining FISH results allowed confirmation of the previously proposed diagnoses.
Figure 1.
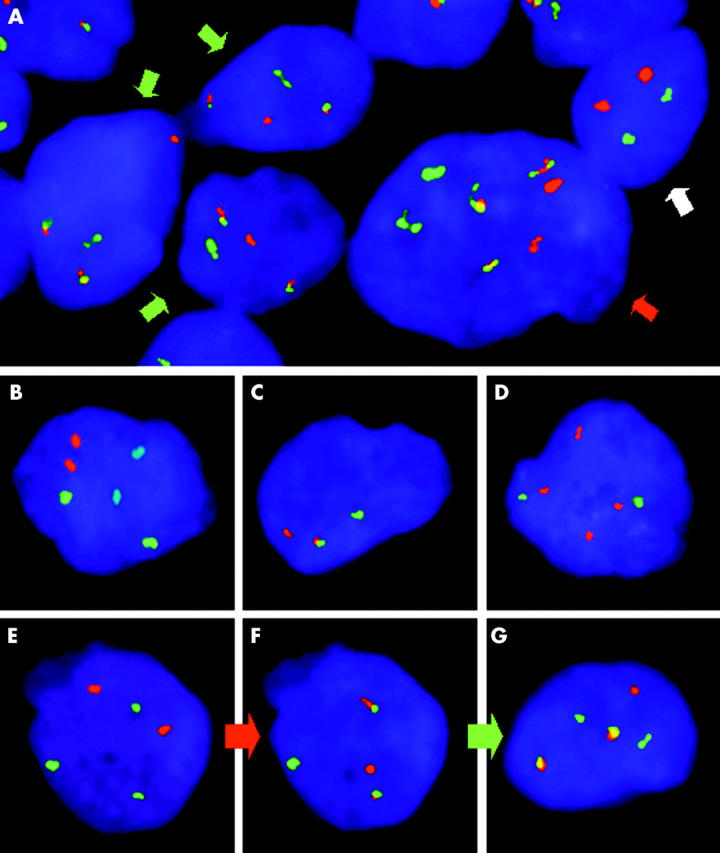
Fluorescence in situ hybridisation (FISH) in tissue imprints. (A) Detection of t(14;18)(q32;q21) in a lymph node imprint from patient number 15. Together with normal cells (2R2G pattern, white arrow), different clones positive for the IGH–BCL2 translocation are present, one of which discloses the typical FISH pattern (1R1G2F, green arrows), whereas another shows an atypical pattern (2R2G3F, red arrow). (B) FISH for t(8;14)(q24;q32) in a lymph node imprint from patient number 11 showing the typical pattern of normal cells (2A2R2G). (C) FISH for t(3;var)(q27;var) in a brain biopsy imprint from patient number 37 showing the typical pattern associated with BCL6 translocation (1R1G1F). (D) FISH for t(14;18)(q32;q21) in a testicle imprint from patient number 36 showing a negative result (absence of IGH–BCL2 rearrangement) but displaying an atypical FISH pattern with two extra BCL2 signals (4R2G). FISH for (E) t(14;18)(q32;q21) and (F, G) t(11;14)(q13;q32) in a bone marrow smear obtained at diagnosis (E, F) and a lymph node imprint at relapse (G) from patient number 7. Rehybridisation of the same slide with different probes (red arrow) allowed the identification of an IGH–CCND1 translocation (F) in a cell previously identified as negative for the IGH–BCL2 rearrangement (E). This case underwent clonal evolution (green arrow) because the sample obtained at relapse showed an atypical FISH pattern (G) with an extra IGH signal (1R2G2F) not present at diagnosis (F).
Translocation t(14;18)
Thirty samples from 28 different patients were studied for IGH–BCL2 translocations. Nineteen samples showed positive results and 11 were negative (table 1). Atypical FISH patterns were seen in seven of the positive (possibly as a result of variant or complex translocations) and five of the negative (with a split IGH signal as a result of a t(11;14) translocation, except for patients 39 and 40, where it was possibly caused by polyploidy). In addition, eight patients (numbers 15, 19, 21, 22, 25, 29, 39, and 40) showed different clones simultaneously. FISH results confirmed the previous diagnosis in all patients.
Translocation t(8;14)
Six samples from five patients were all negative for IGH–MYC fusions. One ascitic fluid cytoprep from patient number 39 showed atypical FISH patterns (possibly as a result of polyploidy) corresponding to two different clones (table 1). In all these cases, FISH ruled out the diagnosis of Burkitt lymphoma.
Translocation t(3;var)
Ten patients negative for t(11;14) and t(14;18) were subsequently studied for rearrangements involving the BCL6 gene (table 1). BCL6 rearrangements were demonstrated in four patients, whereas the remaining six were negative, allowing proper classification. Only one positive sample (patient number 37) showed an atypical FISH pattern, with an extra BCL6 signal.
DISCUSSION
Different authors have successfully performed FISH on tissue imprints or archival BM smears to detect different chromosomal abnormalities such as trisomies8 and HER2-neu genomic amplification.9 Despite the fact that BM smears can be used for this purpose, tissue imprints and cytopreps have not been used routinely in the study of solid and haematological tumours.
“Fluorescence in situ hybridisation can be performed directly on tissue imprints without further manipulation of the samples”
FISH is routinely used for the diagnosis and classification of lymphoproliferative syndromes.1 Commercial probes are currently available to detect many of the chromosomal abnormalities relevant to the pathogenesis of lymphomas. FISH performed on paraffin wax embedded tissue has been used in the diagnosis of mantle cell lymphoma,10,11 follicular lymphoma,12 and Burkitt lymphoma.13 Nevertheless, performing FISH on paraffin wax embedded tissue may have technical difficulties, such as those associated with cell truncation and overlapping, in addition to problems related to fixation and embedding procedures that interfere with DNA hybridisation. Some authors have tried to overcome these problems by different means, including nuclear isolation from paraffin wax embedded tissue.6 However, these are technically challenging and time consuming methods. Our study was not primarily intended to compare results in paraffin wax embedded and cytological material but to show that tissue imprints and cytopreps, in addition to BM smears if these are not available, constitute a suitable complementary approach in the diagnosis of lymphoproliferative syndromes. Cytological samples can be easily and routinely obtained, air dried, and stored under almost any conditions without the need of sophisticated equipment. They can be safely sent by conventional mail to reference laboratories. This falls under the scope of all kinds of laboratories, regardless of technical or geographical constraints. In addition, FISH can be performed directly on tissue imprints without further manipulation of the samples. The FISH protocol is easy and rapid, produces high quality hybridisations, and microscope examination is straightforward. Cell scoring is easier than in dewaxed tissue sections because the cells are in a monolayer and are therefore easy to identify. The results are not hampered by the use of fixatives and reagents necessary for the embedding process. Moreover, tissue imprints, cytopreps, and BM smears require a smaller amount of sample, which can be crucial in some cases. In this context, it should be noted that FISH can also be performed on fine needle aspiration smears.14
FISH produced useful results in all patients studied. Even when there was insufficient signal at the first attempt, in most cases (nine of 10) this was solved after washing and rehybridising the sample with the same probe. A similar approach was also used successfully to test different probes on the same sample in 14 cases in which a single slide was available.
The fact that FISH can be performed successfully on archival tissue imprints, cytopreps, and BM smears15 opens up the possibility of performing retrospective studies to analyse the evolution of the disease. So far, we have retrospectively studied archival BM smears stored at room temperature for as long as 12 years, and good hybridisation signals and clear cut results were obtained. Indeed, in patient number 7 (table 1), this strategy enabled us to reclassify the diagnosis retrospectively. This particular case, which involved t(11;14), also emphasises the power of the technique for the identification of clonal evolution (fig 1).
In most cases (45 of 46), FISH results confirmed the previously proposed diagnosis. Not only positive results but also negative results can be useful for reaching a proper diagnosis, as in case 38, which was negative for BCL2 and BCL6 rearrangements but, after revisiting previously available data, was diagnosed as a marginal zone lymphoma. In spite of this, negative results do not necessarily preclude a previously suspected diagnosis, as exemplified by patient 16, in whom the original diagnosis (follicular lymphoma) was maintained even after negative results for the above rearrangements.
Take home messages.
Fluorescence in situ hybridisation using tissue imprints, cytopreps, and bone marrow smears is a rapid and useful ancillary approach for detecting chromosomal abnormalities in lymphoproliferative syndromes
This technique could be used on a routine basis whenever fresh samples are available
FISH results are also useful for classifying lymphomas in the human immunodeficiency virus setting, as in patient 35, in whom the diagnosis of Burkitt lymphoma was discarded after negative results for t(8;14).
In summary, our study clearly shows that FISH using tissue imprints, cytopreps, and BM smears is an easy, rapid, and reliable method to detect chromosomal abnormalities with high sensitivity and specificity in lymphoproliferative syndromes. Therefore, we suggest that tissue imprints should routinely be taken for FISH analysis, whenever fresh sample is available.
Acknowledgments
The authors are indebted to A Simón and G González-Pardo for their excellent technical assistance. This work was partially supported by the Fondo de Investigaciones Sanitarias (Spain; grants 01/3035 to IB, 02/1177 to JLDM and G03/179 to JM).
Abbreviations
BM, bone marrow
FISH, fluorescence in situ hybridisation
SSC, saline sodium citrate
REFERENCES
- 1.Jaffe ES, Harris NL, Stein H, et al., eds. Pathology and genetics of tumours of haematopoietic and lymphoid tissues. World Health Organisation classification of tumours. Lyon: IARC Press, 2001.
- 2.Ince C, Blick M, Lee M, et al. Bcl-1 gene rearrangements in B-cell lymphoma. Leukemia 1998;2:343–6. [PubMed] [Google Scholar]
- 3.Tsujimoto Y, Finger LR, Yunis J, et al. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science 1984;226:1097–9. [DOI] [PubMed] [Google Scholar]
- 4.Croce CM, Nowell PC. Molecular basis of human B cell neoplasia. Blood 1985;65:1–7. [PubMed] [Google Scholar]
- 5.Tibiletti MG, Bernasconi B, Dionigi A, et al. The applications of FISH in tumor pathology. Adv Clin Pathol 1999;3:111–18. [PubMed] [Google Scholar]
- 6.Paternoster SF, Brockman SR, McClure RF, et al. A new method to extract nuclei from paraffin-embedded tissue to study lymphomas using interphase fluorescence in situ hybridization. Am J Pathol 2002;160:1967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buño I, Moreno-López E, Díez-Martín JL. Sequential fluorescence in situ hybridization for the quantification of minimal residual disease in recipient cells after sex-mismatched allogeneic stem cell transplantation. Br J Haematol 2002;118:349. [DOI] [PubMed] [Google Scholar]
- 8.Dubinsky R, Amiel A, Manor Y, et al. Fluorescence in situ hybridization (FISH) for retrospective detection of trisomies 3 and 7 in multiple myeloma. Cancer Genet Cytogenet 1995;83:115–18. [DOI] [PubMed] [Google Scholar]
- 9.Moore JG, To V, Patel SJ, et al. HER2/neu gene amplification in breast imprint cytology analyzed by fluorescence in situ hybridization: direct comparison with companion tissue sections. Diagn Cytopathol 2000;23:299–302. [DOI] [PubMed] [Google Scholar]
- 10.Remstein ED, Kurtin PJ, Buño I, et al. Diagnostic utility of fluorescence in situ hybridization in mantle-cell lymphoma. Br J Haematol 2000;110:856–62. [DOI] [PubMed] [Google Scholar]
- 11.Sun T, Nordberg ML, Cotelingam JD, et al. Fluorescence in situ hybridization: method of choice for a definitive diagnosis of mantle cell lymphoma. Am J Hematol 2003;74:78–84. [DOI] [PubMed] [Google Scholar]
- 12.Hirose Y, Masaki Y, Ozaki M. Fluorescence in situ hybridization detection of chromosome IGH/BCL2 translocation from paraffin-embedded tissue: evaluation in follicular lymphoma. Int J Hematol 2003;78:154–9. [DOI] [PubMed] [Google Scholar]
- 13.Haralambieva E, Banham AH, Bastard C, et al. Detection by the fluorescence in situ hybridization technique of MYC translocations in paraffin-embedded lymphoma biopsy samples. Br J Haematol 2003;121:49–56. [DOI] [PubMed] [Google Scholar]
- 14.Bentz JS, Rowe LR, Anderson SR, et al. Rapid detection of the t(11;14) translocation in mantle cell lymphoma by interphase fluorescence in situ hybridization on archival cytopathologic material. Cancer 2004;102:124–31. [DOI] [PubMed] [Google Scholar]
- 15.Hammond DW, Hinchiffle RF, Goyns MH, et al. Application of fluorescence in situ hybridization to chromosome analysis of aged bone marrow smears. J Clin Pathol 1994;47:508–11. [DOI] [PMC free article] [PubMed] [Google Scholar]


