Abstract
Background: There are now several lines of evidence to suggest that protein synthesis and translation factors are involved in the regulation of cell proliferation and cancer development.
Aims: To investigate gene expression patterns of eukaryotic releasing factor 3 (eRF3) in gastric cancer.
Methods: RNA was prepared from 25 gastric tumour biopsies and adjacent non-neoplastic mucosa. Real time TaqMan reverse transcription polymerase chain reaction (RT-PCR) was performed to measure the relative gene expression levels. DNA was isolated from tumour and normal tissues and gene dosage was determined by a quantitative real time PCR using SYBR Green dye.
Results: Different histological types of gastric tumours were analysed and nine of the 25 tumours revealed eRF3/GSPT1 overexpression; moreover, eight of the 12 intestinal type carcinomas analysed overexpressed the gene, whereas eRF3/GSPT1 was overexpressed in only one of the 10 diffuse type carcinomas (Kruskal-Wallis Test; p < 0.05). No correlation was found between ploidy and transcript expression levels of eRF3/GSPT1. Overexpression of eRF3/GSPT1 was not associated with increased translation rates because the upregulation of eRF3/GSPT1 did not correlate with increased eRF1 levels.
Conclusions: Overexpression of eRF3/GSPT1 in intestinal type gastric tumours may lead to an increase in the translation efficiency of specific oncogenic transcripts. Alternatively, eRF3/GSPT1 may be involved in tumorigenesis as a result of its non-translational roles, namely (dis)regulating the cell cycle, apoptosis, or transcription.
Keywords: gastric cancer, eRF3/GSPT1, gene expression, real time polymerase chain reaction, translation machinery
Gastric cancer is a major cause of cancer related death worldwide, particularly in developing countries.1 Two histological types of tumour are recognised according to Laurén’s classification: the intestinal and the diffuse types, which are characterised by different carcinogenic pathways.2–4 Frequent genomic aberrations,5–7 differential expression of some genes,8,9,10 genomic instability,11,12 and specific mutations13–15 have been reported in gastric carcinomas, but the precise molecular events underlying the development of gastric tumours are still not clear. At present, prognosis remains poor: most patients have advanced disease at diagnosis and curative surgery remains the most effective treatment.16
“Several lines of evidence support a role for eukaryotic translation factors in cancer development, but the role they play in the development and progression of cancer is still unknown”
The involvement of protein synthesis in the regulation of cell proliferation and cancer development is currently one important field in cancer research.17–21 Changes in the expression pattern of translation factors can lead to several changes in the tumour cell, such as an increase in the overall rate of protein synthesis and/or overexpression of specific proteins involved in cell growth and proliferation.21–24 Several lines of evidence support a role for eukaryotic translation factors in cancer development, but the role they play in the development and progression of cancer is still unknown. Several translation initiation and elongation factors have been shown to be overexpressed in different types of cancers—for example, eukaryotic initiation factor 3S8 (eIF3S8; 16p11.2) in testicular seminomas,25 eIF3-p40 (8q23) in breast and prostate cancers,26 eIF-4E (4q21–q25) and eIF-2α (14q24.1) in non-Hodgkin lymphomas,27 and eEF1A2 (20q13.3) in ovarian tumours28—but there have been no such studies on translation release factors.
Eukaryotic release factor 3 (eRF3) has many functions in eukaryotic cells. It controls the regulation of the cell cycle at the G1 to S phase transition,29 and it regulates protein synthesis as a GTP dependent stimulator of eRF1 in translation termination.30 It was also reported to have a key role as an initiator of the mRNA degradation machinery,31,32 in the recycling of ribosomes in successive cycles of translation,33 and probably also in transcription regulation.34 Two genes encoding eRF3 are presently known, named eRF3/GSPT1 and eRF3/GSPT2, and mapping in the human genome to 16p13.1 and Xp11.21–23, respectively.29,35
Because no studies have examined the expression pattern and the occurrence of genetic alterations of eukaryotic release factors in human tumours, we decided to investigate the role of eRF3/GSPT1 in gastric cancer. In our study, we analysed the expression pattern of the translation termination factors in gastric tumour samples and corresponding non-neoplastic adjacent mucosa using a quantitative real time reverse transcription polymerase chain reaction (RT-PCR). We also determined eRF3/GSPT1 gene dosage by real time PCR in 35 gastric tumours.
MATERIALS AND METHODS
Patients and tissue samples
We analysed 35 gastric tumours collected from surgical resections after informed consent was obtained. The tumours were diagnosed as gastric cancers after histopathological examination performed in the department of pathology. Patients were aged 33–87 years (mean, 66.3).
Tissue samples were stored in RNAlater (Ambion, Austin, Texas, USA) at −20°C immediately after surgery until RNA extraction. Freshly frozen samples from 25 gastric tumours were obtained, together with a sample from the adjacent non-cancerous tissue, for gene expression analysis. The samples were classified as 10 diffuse type, 12 intestinal type, and three other carcinomas (one mixed and two early). Ten additional formalin fixed, paraffin wax embedded carcinoma samples and adjacent normal tissues were used for the gene dosage assay.
DNA extraction, RNA extraction, and cDNA synthesis
RNA was extracted from fresh tissues using the SV Total RNA Isolation System (Promega, Mannheim, Germany), as recommended by the manufacturer. First strand cDNA was synthesised using MultiScribe™ reverse transcriptase (Applied BioSystems, Foster City, California, USA), with random hexamers, according to the manufacturer’s instructions in a total volume of 50 μl; samples were incubated for 10 minutes at 25°C, 30 minutes at 48°C, and finally for five minutes at 95°C.
DNA was extracted from two 7 μm thick formalin fixed, paraffin wax embedded tissue sections using chelex resins36 and from fresh tissues using the DNeasy tissue kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol.
Real time PCR
Gene expression levels
Expression of the eukaryotic translation factors eRF3/GSPT1, eRF3/GSPT2, and eRF1 in tumour samples relative to their normal adjacent tissues was investigated using real time quantitative RT-PCR based on TaqMan® fluorescence methodology. Gene specific primers and TaqMan probes labelled with 5′FAM and 3′TAMRA were designed for each gene (table 1). To avoid the detection of amplification of genomic DNA the probes were localised in an exon–exon boundary (except for eRF3/GSPT2, which does not have introns). Human 18S rRNA (TaqMan predeveloped assay reagents for gene expression; Applied BioSystems) was used as the endogenous control gene. Reactions were performed with TaqMan Universal PCR Master Mix (Applied BioSystems) in a 25 μl reaction volume. All reactions were performed in triplicate and included a negative control. PCR reactions were performed in the GeneAmp® 5700 Sequence Detection System (Applied BioSystems). Cycling conditions were: two minutes at 50°C, 10 minutes at 95°C, and 40 cycles of 15 seconds at 95°C and one minute at 60°C.
Table 1.
Primers and probes used in real time PCR assays
| Oligonucleotide sequence (5′–3′) | |
| Gene expression | |
| eRF1 | F: TGCATCTAACATTAAGTCACGAGT |
| R: TCCACAGTATACAACCAGACCATT | |
| P: AACCGCCTTTCAGTCCTGGGAGCC | |
| eRF3/GSPT1 | F: CGCCAGGTGCTCCTAAGAAAG |
| R: CAAATACATTATTTGTCCTCCAATGGT | |
| P: ACTTGCCAGCATCTACGTGCCCAATG | |
| eRF3/GSPT2 | F: GAGTTCGTGCCGTCCTTCCT |
| R: CCCGACTCAGCCGCCCACCCT | |
| P: CCGTCCCATCCTTTTACCTTGAG | |
| Gene dosage | |
| eRF3/GSPT1 | F: ATAATGATGTACACGGTCACAGTT |
| R: TTCTTGAAGAGAGATTAGTATGATGAA | |
| β Actin | F: AACACTGGCTCGTGTGACAA |
| R: CAGACCTACTGTGCGCCTACT |
Primers and TaqMan probes used in gene expression assays were designed in Molecular Baecon software (BioRad); primers used in the gene dosage assay were designed in Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi).
eRF, eukaryotic release factor; F, forward primer; P, TaqMan probe; PCR, polymerase chain reaction; R, reverse primer.
Relative quantification of the mRNA levels of the target genes (quantity of transcripts of the target in tumour samples relative to normal tissues) was determined using the ΔΔCT method. Briefly, the amount of target was normalised to the endogenous reference gene (18S rRNA) and its expression in tumour samples was calculated relative to a calibrator (normal adjacent sample). Final results are expressed as N-fold difference in tumour expression relative to non-cancerous adjacent tissue.
Gene dosage of eRF3/GSPT1
Gene dosage was assessed by quantitative real time PCR using SYBR Green dye I as the fluorescent signal. This assay relies on a comparison of the amount of product generated from the target gene (eRF3/GSPT1) and that generated from a disomic reference gene (β actin), assuming equal PCR efficiencies for both. Table 1 shows the primers used for eRF3/GSPT1 and β actin genomic DNA. A fragment of genomic DNA from eRF3/GSPT1 was amplified using SYBR Green PCR Master Mix (Applied BioSystems) in a 20 μl reaction volume and the endogenous control gene β actin was measured simultaneously. Each series of PCR reactions included triplicates for all tumour and normal samples, a non-template control, and a four point standard curve, established using serial dilutions (1/2, 1/5, 1/10, and 1/100) of a known disomic sample for both genes. PCR reactions were performed in the iCycler iQ Real-Time PCR Detection System (BioRad, Hercules, California, USA). Cycling conditions were: two minutes at 50°C, 10 minutes at 95°C, and 40 cycles of 15 seconds at 95°C and one minute at 60°C. To confirm the specificity of the PCR, after the amplification reaction, the products were submitted to a melting curve analysis and subsequent gel electrophoresis.
In each experiment the threshold cycle was manually adjusted and Ct values were averaged for each sample. A standard curve was established for each gene using the serial dilutions. For each sample, the amount of eRF3/GSPT1 and β actin was determined from the standard curve. The gene dosage of eRF3/GSPT1 was calculated by the ratio of its amount and the amount of the reference gene. Theoretically, the ratio of tumour to normal is expected to be 1 for disomic tumour samples, greater than 1 if there are allele amplifications, and less than 1 in the presence of allele deletions.
Statistical analysis
The relation of gene expression levels and histological types of gastric tumours was evaluated by a Kruskal-Wallis H test performed in SPSS 12.0 for Windows. Differences were considered significant when the p values were less than 0.05.
RESULTS
Gene expression levels
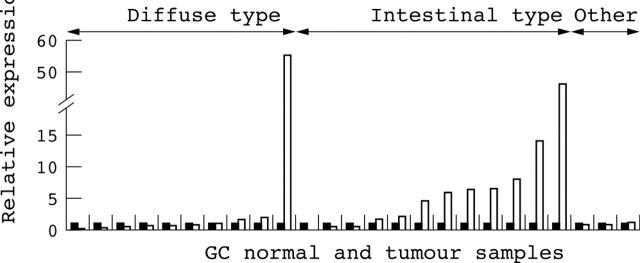
Nine of the 25 gastric cancers examined overexpressed eRF3/GSPT1 in tumour tissue compared with adjacent non-neoplastic mucosa (fig 1). To evaluate the relation between the level of eRF3/GSPT1 expression and the different histopathological types, which differ in their epidemiology, aetiology, pathogenesis, and behaviour,2–4 tumours were classified into groups as follows: intestinal type carcinomas (well differentiated cells, which retain cell cohesion, allowing the formation of glandular structures with sharp margins), diffuse type carcinomas (contain small undifferentiated cells or small cell clusters with deceptive margins, which invade large areas of the stomach and can lead to early metastasis), and other types, including mixed and early carcinomas. Overexpression was seen in eight of the 12 intestinal type carcinomas analysed, but in only one of the 10 diffuse type carcinomas (table 2).
Figure 1.
Relative expression of eukaryotic release factor 3/GSPT1 in 25 gastric cancer (GC) tumours (open bars) and normal adjacent mucosa (black bars) analysed by TaqMan real time reverse transcription polymerase chain reaction.
Table 2.
Frequency of tumours in which eRF3/GSPT1 was underexpressed, overexpressed, or did not vary
| Histological classification | Underexpressed | No variation | Overexpressed |
| Diffuse | 3/10 (30%) | 6/10 (60%) | 1/10 (10%) |
| Intestinal | 1/12 (8.3%) | 3/12 (25%) | 8/12 (66.7%) |
| Others | 0/3 (0%) | 3/3 (100%) | 0/3 (0%) |
Expression was measured by real time polymerase chain reaction.
Differences in overexpression and underexpression between the histological types were assessed using the Kruskal-Wallis test and p<0.05 was taken as significant.
Relative overexpression was detected in nine of 25 tumours when compared with non-neoplastic mucosa. Different levels of expression were detected: in two tumours, expression was two to fivefold higher; in four tumours it was five to tenfold higher; and in three it was more than tenfold higher than in normal adjacent mucosa. In only four of 25 tumours was expression less than half that seen in non-neoplastic mucosa and in none was it more than five times less.
Only one of 10 diffuse type carcinomas overexpressed the gene, whereas overexpression was seen in eight of 12 intestinal type tumours; none of the other types of tumours overexpressed or underexpressed eRF3/GSPT1.
To investigate whether there was a correlation between the eRF3/GSPT1 expression level and translation termination, and consequently translation rates, we analysed the expression pattern of the gene encoding eRF1—the most important factor in the termination step of translation. In intestinal type tumours, eRF1 was overexpressed in a third of the samples and underexpressed in another third. In diffuse type cancers, this gene was underexpressed in half of the samples and overexpressed in a quarter. There were no correlations between eRF3/GSPT1 and eRF1 gene expression levels.
The pattern of eRF3/GSPT2 expression was also investigated, but no expression was detected either in normal mucosa or in gastric tumour samples.
eRF3/GSPT1 gene dosage assay
Validation of standard curves
Standard curves were constructed for target and control genes for every assay. The dynamic range was wide. A strong linear correlation between the Ct and the log of the starting amount of DNA was determined (r2 > 0.990).
The efficiency (E) of amplification, calculated by the formula E = 101/m − 1 (where m is the slope of the standard curve), was similar for both genes and ranged from 95% to 100% in all assays.
Melting curves and gel electrophoresis showed the presence of a single band of the expected size.
Gene dosage quantification
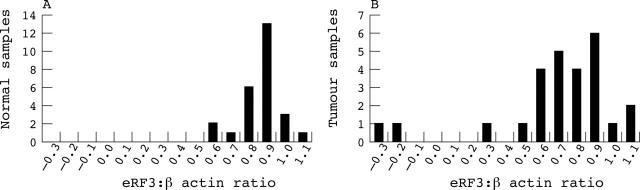
Quantitative results from the normal tissues were first evaluated for gene dosage alterations and ranged from 0.69 to 1.20 (fig 2A), which is in the range of expected values for disomic samples. Loss of genetic material was seen in three tumours (fig 2B). We have not found eRF3/GSPT1 amplification in the samples studied so far.
Figure 2.
Histogram showing distribution of gene dosage obtained by SYBR Green real time polymerase chain reaction in (A) 25 normal gastric mucosal samples and (B) 25 gastric tumours. In the normal samples, eRF3/GSPT1 ranged from 0.69 to 1.20 relative to β actin. Three tumours showed loss of genetic material.
Relation between eRF3/GSPT1 levels of expression and gene dosage
Among the tumours analysed for eRF3/GSPT1 expression we detected allele deletions in three tumours (two intestinal and one diffuse type) and none of them was underexpressed. Thus, there was no association between eRF3/GSPT1 expression and gene copy number.
DISCUSSION
Using real time RT-PCR, we have shown that eRF3/GSPT1, which encodes eukaryotic release factor 3, is highly expressed at the mRNA level in eight of 12 intestinal type gastric tumours, whereas only one in 10 of the diffuse type tumours overexpressed this gene. Several factors can lead to changes in the gene expression patterns seen in tumour cells. One such factor is DNA copy number alterations, which can lead to underexpression or overexpression of certain genes and thus be responsible for cancer development. The detection of deletions (especially monoallelic deletions) in tumour samples is particularly difficult because they are present in only a fraction of the cells in the sample as a result of contamination with adjacent non-cancerous cells. It is also difficult to choose an endogenous control gene to use as reference, because aneuploidy is frequent in tumours. We solved these difficulties by using real time PCR, which is a very accurate and sensitive technique, and using the β actin gene as the disomic endogenous control reference; this gene showed minimal variation among all the normal and tumour samples analysed. Furthermore, tumour samples always showed minimal residual contamination with normal cells and were as homogeneous as possible.
Because we did not detect amplification of eRF3/GSPT1 in the tumour samples analysed for gene dosage, changes in gene copy number cannot be responsible for the overexpression of the gene seen in intestinal type gastric carcinomas, so that alterations in the regulation of gene expression must be involved.
The overexpression of eRF3/GSPT1 was not associated with increased translation rates because the upregulation of eRF3/GSPT1 did not correlate with increased amounts of eRF1. We also investigated the expression of eRF3/GSPT2, but this gene was not expressed in gastric tissues. In addition, we failed to detect eRF3/GSPT2 expression in breast and small intestinal tissues (data not shown). Interestingly, gene expression was detected in thyroid gland and liver tissue, indicating that the expression of this factor has a tissue specific pattern.
The molecular pathway by which eRF3/GSPT1 overexpression contributes to gastric cancer development is unclear, although there are several possible mechanisms.
It is now known that signalling pathways do not stimulate the translation of all mRNAs equally. Changes in the expression or availability of the components of the translation machinery can lead to an increase in the overall rate of protein synthesis or can affect the spectrum of mRNAs translated,21,24,37–39 activating the translation of specific mRNA molecules involved in growth and proliferation, leading to the overexpression of their products.17 Therefore, it is possible that the overexpression of eRF3/GSPT1 could increase the translation efficiency of specific oncogenic transcripts.
Although there are differences in the proliferation rate between intestinal and diffuse type gastric tumours, inconsistent and sometimes conflicting results have been reported. Nevertheless, lower proliferating rates have been assigned to intestinal tumours using different labelling indexes.40,41 Therefore, it seems unlikely that the higher amounts of eRF3/GSPT1 mRNA seen in intestinal type tumours are necessary for their rapid proliferating activity. Moreover, about a third of the samples underexpressed eRF1, the key gene for translation termination, which can result in reduced translation efficiency. However, in diffuse type tumours, half of the samples underexpressed eRF1, and eRF3 levels were not altered.
“The effects of eRF3/GSPT1 on chromosome segregation and cytokinesis could be a possible mechanism by which this gene influences tumorigenesis”
Other non-translational roles of eRF3 have recently been studied. This gene was shown to affect the tubulin cytoskeleton,42 suggesting a role in the control of chromosome segregation. More recently, its involvement in the assembly of the actin cytoskeleton in yeast was also demonstrated.43 Deregulation of eRF3 expression also resulted in abnormal meiotic chromosome segregation and defects in cytoskeleton assembly in spermatids of Drosophila melanogaster.44 Taking all these results into consideration, the effects of eRF3/GSPT1 on chromosome segregation and cytokinesis could be a possible mechanism by which this gene influences tumorigenesis.
Take home messages.
eRF3/GSPT1 was overexpressed in eight of 12 intestinal type gastric tumours
This overexpression may lead to an increase in the translation efficiency of specific oncogenic transcripts
Alternatively, eRF3/GSPT1 may be involved in tumorigenesis as a result of its non-translational roles, namely (dis)regulating the cell cycle, apoptosis, or transcription
A proteolytically processed isoform of the eRF3/GSPT1 protein, which functions as an IAP binding protein (inhibitor of apoptosis), has recently been described.45 This newly identified role of eRF3/GSPT1, which can potentiate or inhibit apoptosis, could be another possible mechanism by which this gene influences cancer development. Different patterns of expression of several apoptosis related proteins have been found in intestinal and diffuse types of gastric tumour. Diffuse type tumour cells may avoid cellular death as a result of the expression of survivin, an apoptosis inhibiting protein,46 and Bcl-xl, an antiapoptotic protein, in addition to the absence of Fas, a member of the tumour necrosis factor receptor family.47 The increased expression of eRF3/GSPT1 in intestinal type tumours might have an effect on the apoptosis pathway.
Our study provides compelling evidence for the involvement of translation factors in tumorigenesis. Moreover, this is the first time that a release factor has been associated with the development or progression of cancer.
Acknowledgments
We thank B Carmona for laboratory assistance and L Gonçalves (Hospital Fernando de Fonseca), MJ Forjaz de Lacerda, and H Bicha Castelo (Hospital de Santa Maria). This work was supported by a grant from Fundação para a Ciência e Tecnologia (MCES) – POCTI/MGI/40071/2001.
Abbreviations
eIF, eukaryotic initiating factor
eRF, eukaryotic release factor
PCR, polymerase chain reaction
RT, reverse transcription
REFERENCES
- 1.Roder DM. The epidemiology of gastric cancer. Gastric Cancer 2000;5:5–11. [DOI] [PubMed] [Google Scholar]
- 2.Stadtlander C, Waterbor J. Molecular epidemiology, pathogenesis and prevention of gastric cancer. Carcinogenesis 1999;20:2195–207. [DOI] [PubMed] [Google Scholar]
- 3.Werner M, Becker KF, Keller G, et al. Gastric adenocarcinoma: pathomorphology and molecular pathology. J Cancer Res Clin Oncol 2001;127:207–16. [DOI] [PubMed] [Google Scholar]
- 4.Yuasa Y. Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat Rev Cancer 2003;3:592–600. [DOI] [PubMed] [Google Scholar]
- 5.Wu CW, Chen GD, Fann CS, et al. Clinical implications of chromosomal abnormalities in gastric adenocarcinomas. Genes Chromosomes Cancer 2002;35:219–31. [DOI] [PubMed] [Google Scholar]
- 6.Varis A, Wolf M, Monni O, et al. Targets of gene amplification and overexpression at 17q in gastric cancer. Cancer Res 2002;62:2625–9. [PubMed] [Google Scholar]
- 7.Molnar B, Bocsi J, Karman J, et al. Immediate DNA ploidy analysis of gastrointestinal biopsies taken by endoscopy using a mechanical dissociation device. Anticancer Res 2003;23:655–60. [PubMed] [Google Scholar]
- 8.El-Rifai W, Smith MF Jr, Li G, et al. Gastric cancers overexpress DARPP-32 and a novel isoform, t-DARPP. Cancer Res 2002;62:4061–64. [PubMed] [Google Scholar]
- 9.Rosivatz E, Becker I, Specht K, et al. Differential expression of the epithelial–mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol 2002;161:1881–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nabais S, Machado JC, Lopes C, et al. Different patterns of beta-catenin expression in gastric carcinomas: relationship with clinicopathological parameters and prognostic outcome. Histopathology 2002;41:368–9. [DOI] [PubMed] [Google Scholar]
- 11.Wu MS, Lee CW, Shun CT, et al. Clinicopathological significance of altered loci of replication error and microsatellite instability-associated mutations in gastric cancer. Cancer Res 1998;58:1494–7. [PubMed] [Google Scholar]
- 12.Conde AR, Martins G, Saraiva C, et al. Association of p53 genomic instability with the glutathione S-transferase null genotype in gastric cancer in the Portuguese population. Mol Pathol 1999;52:131–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huntsman DG, Carneiro F, Lewis FR, et al. Early gastric cancer in young asymptomatic carriers of germ-line E-cadherin mutations. N Engl J Med 2001;344:1904–9. [DOI] [PubMed] [Google Scholar]
- 14.Silva F, Carvalho F, Peixoto A, et al. MUC1 gene polymorphism in the gastric carcinogenesis pathway. Eur J Hum Genet 2001;9:548–52. [DOI] [PubMed] [Google Scholar]
- 15.Frazier ML, Inamdar N, Alvula S, et al. Few point mutations in elongation factor-1gamma gene in gastrointestinal carcinoma. Mol Carcinog 1998;22:9–15. [PubMed] [Google Scholar]
- 16.Alberts SR, Cervantes A, Van De Velde CJ. Gastric cancer: epidemiology, pathology and treatment. Ann Oncol 2003;14:II31–6. [DOI] [PubMed] [Google Scholar]
- 17.Caraglia M, Budillon A, Vitale G, et al. Modulation of molecular mechanisms involved in protein synthesis machinery as a new tool for the control of cell proliferation. Eur J Biochem 2000;267:3919–36. [DOI] [PubMed] [Google Scholar]
- 18.Clemens MJ, Bommer UA. Translational control: the cancer connection. Int J Biochem Cell Biol 1999;31:1–23. [DOI] [PubMed] [Google Scholar]
- 19.Fingar DC, Richardson CJ, Tee AR, et al. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol 2004;24:200–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland EC. Regulation of translation and cancer. Cell Cycle 2004;3:452–5. [PubMed] [Google Scholar]
- 21.Rajasekhar VK, Holland EC. Postgenomic global analysis of translational control induced by oncogenic signaling. Oncogene 2004;23:3248–64. [DOI] [PubMed] [Google Scholar]
- 22.Rhoads RE. Signal transduction pathways that regulate eukaryotic protein synthesis. J Biol Chem 1999;274:30337–40. [DOI] [PubMed] [Google Scholar]
- 23.Meric F, Hunt KK. Translation initiation in cancer: a novel target for therapy. Mol Cancer Ther 2002;1:971–9. [PubMed] [Google Scholar]
- 24.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene 2004;23:3189–99. [DOI] [PubMed] [Google Scholar]
- 25.Rothe M, Ko Y, Albers P, et al. Eukaryotic initiation factor 3 p110 mRNA is overexpressed in testicular seminomas. Am J Pathol 2000;157:1597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nupponen NN, Porkka K, Kakkola L, et al. Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am J Pathol 1999;154:1777–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Rosenwald IB, Hutzler MJ, et al. Expression of the eukaryotic translation initiation factors 4E and 2 in non-Hodgkin’s lymphomas. Am J Pathol 1999;55:247–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anand N, Murthy S, Amann G, et al. Gene encoding protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat Genet 2002;31:301–5. [DOI] [PubMed] [Google Scholar]
- 29.Ozawa K, Murakami Y, Eki T, et al. Mapping of the human GSPT1 gene, a human homolog of the yeast GST1 gene, to chromosomal band 16p13.1. Somat Cell Mol Genet 1992;18:189–94. [DOI] [PubMed] [Google Scholar]
- 30.Zhouravleva G, Frolova L, Le Goff X, et al. Termination of translation in eukaryotes in governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 1995;14:4065–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoshino S, Imai M, Kobayashi T, et al. The eukaryotic polypeptide chain release factor 3 (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with PABP. J Biol Chem 1999;274:16677–80. [DOI] [PubMed] [Google Scholar]
- 32.Hosoda N, Kobayashi T, Uchida N, et al. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J Biol Chem 2003;278:38287–91. [DOI] [PubMed] [Google Scholar]
- 33.Uchida N, Hoshino S, Imataka H, et al. Novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in cap/poly(A)-dependent translation. J Biol Chem 2002;277:50286–92. [DOI] [PubMed] [Google Scholar]
- 34.Hoshino S, Imai M, Mizutani M, et al. Molecular cloning of a novel member of the eukaryotic polypeptide releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J Biol Chem 1998;273:22254–9. [DOI] [PubMed] [Google Scholar]
- 35.Hansen LL, Jakobsen CG, Justesen J. Assignment of the human peptide chain release factor 3 (GSPT2) to Xp11.23→p11.21 and of the distal marker DXS1039 by radiation hybrid mapping. Cytogenet Cell Genet 1999;86:250–1. [DOI] [PubMed] [Google Scholar]
- 36.Coombs N, Gough A, Primrose N. Optimisation of DNA and RNA extraction from archival formalin-fixed tissue. Nucleic Acids Res 1999;27:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhoads RE. Signal transduction pathways that regulate eukaryotic protein synthesis. J Biol Chem 1999;274:30337–40. [DOI] [PubMed] [Google Scholar]
- 38.Kochetov AV, Ischenko IV, Vorobiev DG, et al. Eukaryotic mRNAs encoding abundant and scarce proteins are statistically dissimilar in many structural features. FEBS Lett 1998;440:351–5. [DOI] [PubMed] [Google Scholar]
- 39.Hornstein E, Git A, Braunstein I, et al. The expression of poly(A)-binding protein gene is translationally regulated in a growth-dependent fashion through a 5′-terminal oligopyrimidine tract motif. J Biol Chem 1999;274:1708–14. [DOI] [PubMed] [Google Scholar]
- 40.Shinohara T, Ohshima K, Murayama H, et al. Apoptosis and proliferation in gastric carcinoma: the association with histological type. Histopathology 1996;29:123–9. [DOI] [PubMed] [Google Scholar]
- 41.Seno H, Oshima M, Taniguchi MA, et al. CDX2 expression in the stomach with intestinal metaplasia and intestinal-type cancer: prognostic implications. Int J Oncol 2002;21:769–74. [DOI] [PubMed] [Google Scholar]
- 42.Borhsenius AG, Tchourikova AA, Inge-Vechtomov SG. Recessive mutations in SUP35 and SUP45 genes coding for translation release factors affect chromosome stability in Saccharomyces cerevisiae. Curr Genet 2000;37:285–91. [DOI] [PubMed] [Google Scholar]
- 43.Valouev IA, Kushnirov VV, Ter-Avanesyan MD. Yeast polypeptide chain release factors eRF1 and eRF3 are involved in cytoskeleton organization and cell cycle regulation. Cell Motil Cytoskeleton 2002;52:161–73. [DOI] [PubMed] [Google Scholar]
- 44.Basu J, Williams BC, Li Z, et al. Depletion of a Drosophila homolog of yeast Sup35p disrupts spindle assembly, chromosome segregation, and cytokinesis during male meiosis. Cell Motil Cytoskeleton 1998;39:286–302. [DOI] [PubMed] [Google Scholar]
- 45.Hegde R, Srinivasula SM, Datta P, et al. The polypeptide chain releasing factor GSPT1/eRF3 is proteolytically processed into an IAP-binding protein. J Biol Chem 2003;278:38699–706. [DOI] [PubMed] [Google Scholar]
- 46.Wakana Y, Kasuya K, Katayanagi S, et al. Effect of survivin on cell proliferation and apoptosis in gastric cancer. Oncol Rep 2002;9:1213–18. [PubMed] [Google Scholar]
- 47.van der Woude CJ, Kleibeuker JH, Tiebosch AT, et al. Diffuse and intestinal type gastric carcinomas differ in their expression of apoptosis related proteins. J Clin Pathol 2003;56:699–702. [DOI] [PMC free article] [PubMed] [Google Scholar]