Abstract
Background: Malignant melanoma arising from different body compartments may be associated with differing aetiological factors and clinical behaviour, and may manifest diverse molecular genetic profiles. Although many studies have focused on cutaneous melanoma, little is known of mucosal and other types of melanoma. In particular, malignant melanoma of soft parts is different from other melanomas in many respects, yet manifests a common melanocytic differentiation. Mutation of BRAF is now known to be common in cutaneous melanomas, and raises possible new therapeutic options of anti-RAF treatment for these patients. Few data are available for non-cutaneous melanomas.
Aims: To study the incidence of BRAF and NRAS mutations in melanomas arising in diverse internal organs.
Methods: Fifty one melanomas from various internal organs were investigated for BRAF and NRAS mutation by direct DNA sequencing.
Results: BRAF and NRAS mutations were found in two and five mucosal melanomas arising from the aerodigestive and female genital tracts (n = 36). Their occurrence is mutually exclusive, giving a combined mutation incidence rate of 19.4% in mucosal melanomas. Both BRAF and NRAS mutations were absent in malignant melanoma of soft parts (n = 7). BRAF mutation was also absent in uveal melanoma (n = 6), but was seen in two of five cutaneous melanomas. The incidence of BRAF or combined BRAF/NRAS mutations in all non-cutaneous groups was significantly lower than published rates for cutaneous melanomas.
Conclusion: Each melanoma subtype may have a unique oncogenetic pathway of tumour development, and only a small fraction of non-cutaneous melanomas may benefit from anti-RAF treatment.
Keywords: BRAF, NRAS, mucosal melanoma, malignant melanoma of soft parts
Melanoma arises from the malignant transformation of pigment producing cells, the melanocytes. Melanocytes originate in the neural crest and migrate to the basal layer of the epidermis and the hair follicles during embryogenesis. Apart from the skin, melanocytes are also found in most squamous covered mucous membranes, the uveal tract in the eye, and the leptomeninges of the brain. Thus, malignant melanomas can be classified by their site of origin into cutaneous melanoma, mucosal melanoma, uveal (ocular) melanoma, and leptomeningeal melanoma. Interestingly, malignant melanoma can also arise in sites not known to harbour melanocytes. The most prominent example includes malignant melanoma of soft parts, also known as clear cell sarcoma of tendons and aponeuroses. This tumour is characterised by nests of malignant cells with a morphological and immunohistochemical profile of melanocytic differentiation, arising in tendons or aponeuroses in the extremities of young adults. Despite a common neural crest origin, the different subtypes of melanoma are distinct in their age of onset, aetiology, and biological behaviour. For example, cutaneous melanoma and malignant melanoma of soft parts tend to have an earlier age of onset compared with the other groups. Uveal melanoma tends to spread via the haematogenous route, whereas cutaneous melanoma shows a greater preference for lymphatic metastasis. Cutaneous melanoma is by far the most common and most well studied subtype, constituting 90% of all cases. Whereas the development of most cutaneous melanomas is strongly linked to exposure to ultraviolet (UV) irradiation, the aetiology of non-cutaneous melanoma remains obscure. Moreover, melanomas arising from different sites may be heterogeneous in their molecular pathway of tumour development. For example, chromosomal aberrations such as monosomy of chromosome 3 and gain of 8q are common in uveal but rare in cutaneous melanoma.1,2 Conversely, alteration of the CDKN2 locus in 9p21 is common in cutaneous but rare in uveal melanoma.3 The t(12,22)(q13;q12) translocation, resulting in a fusion gene product combining the 5′ region of the EWS gene and the 3′ region of the ATF1 gene, is found in 80% of malignant melanomas of soft parts, but not in other types of melanoma.4,5 Unfortunately, because of their relatively low incidence, genetic changes remain poorly defined for most non-cutaneous melanomas. With the recent development of pathway specific drug targets, defining these genetic changes in various subtypes of melanoma may have important therapeutic implications.
“Interestingly, malignant melanoma can also arise in sites not known to harbour melanocytes”
It has long been known that activation of the RAS–RAF–MEK–ERK–MAPK (mitogen activated protein kinase) pathway plays an important role in cutaneous melanoma development. This involves an activating mutation of NRAS, which is found in over 20% of cutaneous melanomas.6–8 More recently, our group has described a high frequency of an activating BRAF mutation (more than 60%) in melanoma cell lines and human tumour samples.9 BRAF is a serine/threonine kinase that transduces regulatory signals from RAS through MEK to MAPK in the RAS–RAF–MEK–ERK–MAPK signalling cascade. Subsequent studies by others have confirmed these results,10–13 and have shown that BRAF mutation is also present in a high percentage (82%) of cutaneous melanocytic naevi.12 Over 80% of the mutations involve a single base substitution in the kinase domain (T1799A), leading to a V600E amino acid substitution (nucleotide and codon numbers based on NCBI gene bank nucleotide sequence NT_007914, corresponding to T1796A, V599E in the old gene bank sequence NM_004333). We have previously shown that this mutant BRAF could function in a constitutively active form, with consequent raised kinase activity, and could transform NIH3T3 cells.9 More recently, several studies have indicated that in contrast to cutaneous melanomas, BRAF mutation is rare in uveal melanomas.14–17 Three recent studies have also demonstrated a low incidence of BRAF mutation in melanoma arising from non-hair bearing (glabrous) skin that is relatively protected from UV damage because of the thickened cornified layer, and in melanoma arising from mucosa that is totally sun protected.18–20 Studies of the molecular changes in non-cutaneous melanomas could help to clarify their genetic pathway of development, which may be distinct from cutaneous melanoma. Furthermore, these results may have important therapeutic implications because those with BRAF mutations may benefit from specific treatment using a RAF inhibitor. Here, we analysed a large series of non-cutaneous malignant melanomas arising from various internal organs, including malignant melanoma of soft parts, which has not been studied before, for BRAF and NRAS mutations.
METHODS
Materials
We searched the computerised pathology records of the department of pathology of Queen Mary Hospital and Queen Elizabeth Hospital, Hong Kong from 1993 to 2003. The clinical and pathology records of all patients with a diagnosis of malignant melanoma or malignant melanoma of soft parts (also named clear cell sarcoma of tendon and aponeurosis) were retrieved for review. Patients with a diagnosis of primary non-cutaneous malignant melanoma or metastasis were selected for our present study. All except five cutaneous malignant melanomas or documented metastasis from cutaneous melanoma were excluded. The five cases of cutaneous melanoma or distant metastasis were randomly selected and included as controls for our mutation detection method and potential ethnic differences because most of our patients were of Chinese origin. This study was approved by the ethics committee of the University of Hong Kong.
In total, 56 malignant melanomas from various organ sites were finally selected. Table 1 lists in detail their anatomical site of origin. The histological slides were reviewed and appropriate paraffin wax embedded tissue blocks were retrieved. Sections (6 µm thick) were cut and stained with 0.1% methylene blue. Areas of the malignant melanoma were microdissected under a light microscope for DNA extraction using standard protocols, as described previously.21
Table 1.
Mutations of BRAF and NRAS in various types of melanoma
| Site | Mean age (range) | Sample size | No. of mutated BRAF (%) | Nucleotide | Amino acid | No. of mutated NRAS (%) | Nucleotide | Amino acid |
| Mucosal melanoma | 68.1 (33–95) | 36 | 2 (5.6) | 5 (13.9) | ||||
| Head and neck | ||||||||
| Nasopharynx | 13 | 1 (7.7) | T1799A | V600E | 1 (7.7) | G34T | G12C | |
| Maxilla | 3 | 0 (0) | 0 (0) | |||||
| Hard palate or floor of mouth | 4 | 0 (0) | 1 (25) | A183C | Q61H | |||
| Female genital tract | ||||||||
| Cervix | 1 | 0 (0) | 0 (0) | |||||
| Vagina | 4 | 0 (0) | 1 (25) | C181A | Q61K | |||
| Vulva | 3 | 1 (33.3) | A1742T | N581I | 0 (0) | |||
| Gastrointestinal tract | ||||||||
| Oesophagus | 3 | 0 (0) | 2 (66.7) | G35A A183T | G12D Q61H | |||
| Rectum | 3 | 0 (0) | 0 (0) | |||||
| Anus | 2 | 0 (0) | 0 (0) | |||||
| Melanoma of parenchymal organs | ||||||||
| Brain | 23 | 1 | 0 (0) | 1 (100) | C181A | Q61K | ||
| Ovary | 48 | 1 | 0 (0) | 0 (0) | ||||
| Malignant melanoma of soft parts | 27.7 (17–38) | 7 | 0 (0) | 0 (0) | ||||
| Uveal melanoma | 61 (46–80) | 6 | 0 (0) | 1 (16.7) | A182G | Q61R | ||
| Cutaneous melanoma | 55.6 (40–79) | 5 | 2 (40) | T1799A GT1798–99AA | V600E V600K | 0 (0) |
Mutation detection of BRAF and NRAS
Primers were designed to amplify exons 11 (G loop region) and 15 (activation segment) of BRAF, and exons 1 and 2 of NRAS. The primers for BRAF were the same as those used in our previous study.21 The primer sequences were as follows:
BRAF exon 11F: 5′-TTCTGTTTGGCTTGACTTGACTT-3′
BRAF exon 11R: 5′-ACTTGTCACAATGTCACCACATT-3′
BRAF exon 15F: 5′-TCATAATGCTTGCTCTGATAGGA-3′
BRAF exon 15R: 5′-GGCCAAAAATTTAATCAGTGGA-3′
NRAS exon 1F: 5′- GGTTTCCAACAGGTTCTTGC-3′
NRAS exon 1R: 5′- CACTGGGCCTCACCTCTATG-3′
NRAS exon 2F: 5′- CACACCCCCAGGATTCTTAC-3′
NRAS exon 2R: 5′- TGGCAAATACACAGAGGAAGC-3′.
Polymerase chain reactions (PCRs) were performed in 20 μl reactions containing 0.5μM of each primer, 2.5mM Mg2+, 0.25mM dNTPs, and 1 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, California, USA) in 1× PCR buffer. In total, 40 amplification cycles (95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 45 seconds) were performed with an initial denaturation at 95°C for 10 minutes and a final extension at 72°C for 10 minutes. The PCR products were purified with 1 μl ExoI/SAP (37°C for 15 minutes, then 85°C for 15 minutes) and were then sequenced directly on both strands using the BigDye® Terminator v1.1 cycle sequencing kit (Applied Biosystems) according to manufacturer’s protocol and analysed by the Applied Biosystems 3730 DNA analyser. All mutations were re-confirmed by independent PCR reactions and direct sequencing.
Statistical analysis
The differences in incidence of BRAF mutations or combined BRAF/NRAS mutations in our series of non-cutaneous melanomas (malignant melanoma of soft parts, mucosal melanoma, and uveal melanoma) were analysed in comparison with the cumulative incidence from all major published series of primary cutaneous malignant melanomas using Fisher’s exact test.
RESULTS
Table 1 summarises the incidence of BRAF and NRAS mutations in the various types of mucosal, non-cutaneous, and cutaneous malignant melanomas. In total, there were 36 cases of mucosal melanoma, seven cases of malignant melanoma of soft parts (clear cell sarcoma of tendon and aponeurosis), six cases of uveal melanoma, and five cases of cutaneous melanoma. One case of malignant melanoma arising from melanocytes in a mature cystic teratoma of the ovary, and one case of primary melanoma arising in the brain associated with a focus of leptomeningeal melanosis were also included. These cover a comprehensive spectrum of malignant melanomas from the whole body system.
In total, two BRAF mutations were identified in the 36 malignant melanomas arising from the mucosa of the aerodigestive and female genital tract. One mutation was T1799A (V600E) and occurred in a malignant melanoma of the nasopharynx. The other mutation was a novel mutation A1742T (N581I) identified in a vulval malignant melanoma. No BRAF mutation was identified in the seven malignant melanomas of soft parts, six uveal melanomas, or the two melanomas from the ovary and brain. Figure 1 shows representative sequence chromatograms.
Figure 1.
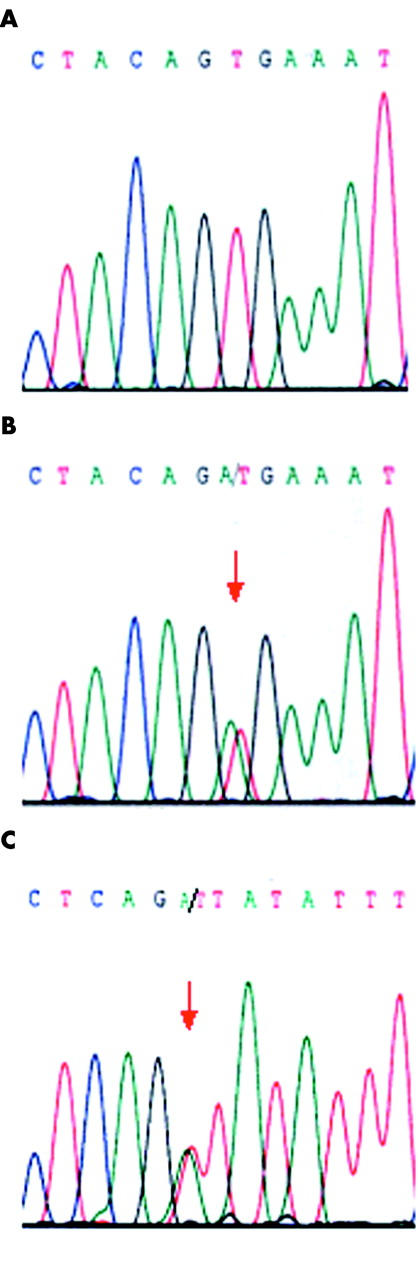
Sequence chromatograms showing (A) wild-type sequence in codon 600 in a malignant melanoma of soft parts; (B) the T1799A (V600E) (arrow) mutation in a malignant melanoma of the nasopharynx; and (C) the A1742T (N581I) (arrow) mutation in a malignant melanoma of the vulva.
Five NRAS mutations were identified in the 36 mucosal melanomas, two involving codon 12 and three involving codon 61. These were from tumours arising from the oesophagus (two cases), nasopharynx (one case), hard palate (one case), or vagina (one case). Two additional NRAS mutations were noted in a uveal melanoma and a melanoma from the brain. None of the malignant melanomas of soft parts harboured an NRAS mutation.
Two BRAF mutations were detected in the five cutaneous melanomas. One mutation, T1799A (V600E), was found in a superficial spreading type melanoma, and the other was a tandem mutation, GT1798-99AA (V600K), occurring in a brain metastasis from a cutaneous melanoma. The three cutaneous melanomas without BRAF mutations were of the acral lentiginous type, the lentigo maligna type, and a melanoma arising from a congenital naevus. No NRAS mutation was detected in the five cutaneous melanomas.
Overall, none of the samples that carried a BRAF mutation had an NRAS mutation.
Table 2 summarises the statistical analysis of differences in incidence of BRAF or combined BRAF/NRAS mutations between the non-cutaneous melanomas from our current series and published series of primary cutaneous melanomas. Both the incidence of BRAF mutation and the combined incidence of BRAF/NRAS mutations were significantly lower in malignant melanoma of soft parts, mucosal melanoma, uveal melanoma, and non-cutaneous melanoma as a group compared with the cutaneous melanomas.
Table 2.
Differences in incidence of BRAF or BRAF/NRAS mutations in various types of non-cutaneous melanomas (present study) compared with primary cutaneous melanomas (from the literature)
| Melanoma type | Incidence of BRAF mutation | p Value* | Combined incidence of BRAF/NRAS mutations | p Value* |
| Primary cutaneous melanoma | 178/444 (40.1%)10,12,15,18,20,22–28† | 111/141 (78.7%)10,22–24,28† | ||
| Malignant melanoma of soft parts | 0/7 (0%) | 0.029 | 0/7 (0%) | <0.0001 |
| Mucosal melanomas | 2/36 (5.6%) | <0.0001 | 7/36 (19.4%) | <0.0001 |
| Uveal melanomas | 0/6 (0%) | 0.048 | 1/6 (16.7%) | 0.0029 |
| All non-cutaneous melanomas | 2/51 (3.9%) | <0.0001 | 9/51 (17.6%) | <0.0001 |
*Comparison of each category against primary cutaneous melanomas using Fisher’s exact test.
†Literature data were pooled from the references.
DISCUSSION
We have identified a BRAF mutation in two of 36 mucosal melanomas arising from the aerodigestive and female genital tracts. Maldonado et al previously studied 21 mucosal melanomas and identified two BRAF mutations.20 Cohen et al studied 25 mucosal melanomas and found one BRAF mutation,18 whereas Edwards et al studied 13 mucosal melanomas and found no BRAF mutations.19 Our findings corroborate those of others that BRAF mutation is rare in mucosal melanomas. Although one of the BRAF mutations seen in the mucosal melanomas involved a previously described amino acid substitution in V600 (previously V599), we identified a novel BRAF mutation, N581I, in a vulval melanoma. Mutation involving the same codon but with a different amino acid substitution (N581S, previously N580S) has been reported in a single case of colorectal cancer.29 The pathogenic relevance of this novel BRAF mutation awaits further functional characterisation.
NRAS mutation was found in five of 36 mucosal melanomas. This is also comparable to a previous study, which recorded NRAS mutations in four of 28 mucosal melanomas.30 In our present study, BRAF and NRAS mutations were mutually exclusive; thus, overall seven of the 36 mucosal melanomas had activation of the RAS–RAF–MEK–ERK–MAPK pathway through either BRAF or NRAS mutation. To our knowledge, this is the first comprehensive study that has characterised both BRAF and NRAS mutations in mucosal melanomas. The combined incidence of BRAF and NRAS mutations is significantly lower (p < 0.0001) than that seen in cutaneous melanomas (table 2).
We found a complete absence of BRAF or NRAS mutations in the seven malignant melanomas of soft parts, which is significantly different from cutaneous melanomas (p < 0.0001). This is the first systematic study of BRAF and NRAS mutation for this type of melanoma. Malignant melanoma of soft parts constitutes a distinct subgroup of malignant melanoma with an intriguing histiogenesis and molecular pathway of tumour development.4,5 These tumours tend to occur in deep tendons and aponeuroses, where melanocytes are not known to exist normally, and there is no involvement of the overlying skin. They have the same immunohistochemical profile as cutaneous melanomas—expressing the melanocytic markers HMB45 and S100 protein—the latter highlighting their neural crest origin. Intracellular melanin, although scanty, can be demonstrated in more than 50% of cases. The prognosis is generally poor, with a high propensity for recurrence and metastasis. Recurrence may sometimes occur after a long latent period of 10 or more years. The characteristic genetic change in these tumours is the t(12;22)(q13;q12) translocation, resulting in the expression of a fusion EWS–ATF1 transcript, which has not been found in cutaneous melanoma. In this respect, translocation involving the EWS gene is also involved in the development of several sarcomas, including Ewing’s sarcoma and primitive neuroectodermal tumour. BRAF mutations are extremely rare in sarcoma, and were seen in only one of 182 (0.5%) cases in our previous mutation screening.9 The rarity of BRAF or NRAS mutations in malignant melanoma of soft parts and the common occurrence of translocation involving the EWS gene suggest that this tumour is more closely related to sarcoma than cutaneous melanoma in its oncogenetic pathway. Interestingly, both BRAF and the EWS–ATF1 fusion gene are involved at different points along the cAMP responsive pathway, which plays an important role in regulating the differentiation and melanogenesis of melanocytes.31–35 Whereas BRAF mediates the cAMP dependent activation of ERKs in melanocytes,36 the EWS–ATF1 chimaeric protein can function as a potent constitutive activator of several cAMP inducible promoters.37,38 It would be interesting to study whether the EWS–ATF1 fusion gene product and mutant BRAF have similar biological roles in melanoma oncogenesis, which could help to explain the absence of BRAF mutations in malignant melanoma of soft parts.
“The difference in incidence of BRAF mutations in different subtypes of melanoma could be related to aetiological factors such as ultraviolet exposure”
The difference in the incidence of BRAF mutation among different subtypes of malignant melanomas is intriguing. BRAF mutation appears to play an important role in regulating the growth of melanocytes and malignant transformation in the cutaneous compartment. Up to 80% of benign naevi of various histological types, including compound naevi, junctional naevi, and congenital naevi, harbour BRAF mutations.12 However, it should be noted that most benign naevi are end stage lesions and do not progress to malignant melanoma. In contrast, a study of cutaneous melanoma revealed a low incidence of BRAF mutation (10%) at the radial growth phase, but a high incidence (63%) in the vertical growth phase, which suggests that BRAF mutation does not occur at the initiation of tumorigenesis but is important in the progression of cutaneous melanoma.10 It has been suggested that the difference in incidence of BRAF mutations in different subtypes of melanoma could be related to aetiological factors such as UV exposure. Accumulating data, including those from the current series, show that the highest incidence of BRAF mutation is found in melanoma that develops in skin subjected to intermittent sun exposure.20 Melanoma developing in skin with chronic sun damage, in glabrous skin shielded from UV by the cornified layer, and in mucosa totally protected from UV exposure generally has a low incidence of BRAF mutation, in the range of 0–15%.18–20 Uveal melanoma and, in this series, melanoma of soft parts do not show BRAF mutation at all.14–17 In contrast, a higher incidence of NRAS mutation was seen in melanoma arising from skin with continuous sun exposure than in skin with intermittent sun exposure or sun protected mucosa.6,8,30,39 Although the mutation spectrum in NRAS in many of the cutaneous melanomas is compatible with UV induced mutations—with a hotspot mutation site in codon 61 involving a TT pyrimidine doublet and its 3′ nucleotide—the most frequent hotspot mutation of BRAF involves a T to A mutation, which is not typical of UV induced mutation. Moreover, a similar mutation spectrum for NRAS genes was found in some of the melanomas arising from internal organs, which are clearly not related to UV exposure. Thus, other environmental or intrinsic biological factors should also be considered as potential causative agents for these mutations in melanomas.
Take home messages.
In contrast to cutaneous melanoma, we found a low incidence of BRAF and NRAS mutations in mucosal melanoma arising from the aerodigestive and female genital tracts, and a complete lack of these mutations in malignant melanoma of soft parts
Each subtype of melanoma may have its own unique genetic pathway of development, and only a small proportion of patients with non-cutaneous melanoma may benefit from anti-RAF specific treatment
In summary, we have documented a low incidence of BRAF and NRAS mutation in mucosal melanoma arising from the aerodigestive and female genital tracts. Moreover, BRAF and NRAS mutation are totally absent in malignant melanoma of soft parts. These data suggest that each subtype of melanoma may have its own unique genetic pathway of development, and only a small proportion of patients with non-cutaneous melanoma may benefit from anti-RAF specific treatment.
Acknowledgments
This work was supported by the Committee on Research and Conference Grant 10205043 from the University of Hong Kong and the Wellcome Trust.
Abbreviations
MAPK, mitogen activated protein kinase
PCR, polymerase chain reaction
UV, ultraviolet
Competing interest: Members of the Cancer Genome Project are inventors of a patent relating to the development of BRAF for treatment and diagnostics
REFERENCES
- 1.Pollock PM, Trent JM. The genetics of cutaneous melanoma. Clin Lab Med 2000;20:667–90. [PubMed] [Google Scholar]
- 2.Singh AD, Wang MX, Donoso LA, et al. Genetic aspects of uveal melanoma: a brief review. Semin Oncol 1996;23:768–72. [PubMed] [Google Scholar]
- 3.Hearle N, Damato BE, Humphreys J, et al. Contribution of germline mutations in BRCA2, P16(INK4A), P14(ARF) and P15 to uveal melanoma. Invest Ophthalmol Vis Sci 2003;44:458–62. [DOI] [PubMed] [Google Scholar]
- 4.Graadt van Roggen JF, Mooi WJ, Hogendoorn PC. Clear cell sarcoma of tendons and aponeuroses (malignant melanoma of soft parts) and cutaneous melanoma: exploring the histogenetic relationship between these two clinicopathological entities. J Pathol 1998;186:3–7. [DOI] [PubMed] [Google Scholar]
- 5.Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: clear cell sarcoma (malignant melanoma of soft parts). Cancer Genet Cytogenet 2001;130:1–7. [DOI] [PubMed] [Google Scholar]
- 6.van’t Veer LJ, Burgering BM, Versteeg R, et al. N-ras mutations in human cutaneous melanoma from sun-exposed body sites. Mol Cell Biol 1989;9:3114–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albino AP, Nanus DM, Mentle IR, et al. Analysis of ras oncogenes in malignant melanoma and precursor lesions: correlation of point mutations with differentiation phenotype. Oncogene 1989;4:1363–74. [PubMed] [Google Scholar]
- 8.Ball NJ, Yohn JJ, Morelli JG, et al. Ras mutations in human melanoma: a marker of malignant progression. J Invest Dermatol 1994;102:285–90. [DOI] [PubMed] [Google Scholar]
- 9.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54. [DOI] [PubMed] [Google Scholar]
- 10.Dong J, Phelps RG, Qiao R, et al. BRAF oncogenic mutations correlate with progression rather than initiation of human melanoma. Cancer Res 2003;63:3883–5. [PubMed] [Google Scholar]
- 11.Gorden A, Osman I, Gai W, et al. Analysis of BRAF and N-RAS mutations in metastatic melanoma tissues. Cancer Res 2003;63:3955–7. [PubMed] [Google Scholar]
- 12.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet 2003;33:19–20. [DOI] [PubMed] [Google Scholar]
- 13.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res 2002;62:6997–7000. [PubMed] [Google Scholar]
- 14.Cohen Y, Goldenberg-Cohen N, Parrella P, et al. Lack of BRAF mutation in primary uveal melanoma. Invest Ophthalmol Vis Sci 2003;44:2876–8. [DOI] [PubMed] [Google Scholar]
- 15.Cruz F III, Rubin BP, Wilson D, et al. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res 2003;63:5761–6. [PubMed] [Google Scholar]
- 16.Edmunds SC, Cree IA, Di Nicolantonio F, et al. Absence of BRAF gene mutations in uveal melanomas in contrast to cutaneous melanomas. Br J Cancer 2003;88:1403–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rimoldi D, Salvi S, Lienard D, et al. Lack of BRAF mutations in uveal melanoma. Cancer Res 2003;63:5712–15. [PubMed] [Google Scholar]
- 18.Cohen Y, Rosenbaum E, Begum S, et al. Exon 15 BRAF mutations are uncommon in melanomas arising in nonsun-exposed sites. Clin Cancer Res 2004;10:3444–7. [DOI] [PubMed] [Google Scholar]
- 19.Edwards RH, Ward MR, Wu H, et al. Absence of BRAF mutations in UV-protected mucosal melanomas. J Med Genet 2004;41:270–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst 2003;95:1878–90. [DOI] [PubMed] [Google Scholar]
- 21.Chan TL, Zhao W, Leung SY, et al. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res 2003;63:4878–81. [PubMed] [Google Scholar]
- 22.Kumar R, Angelini S, Hemminki K. Activating BRAF and N-Ras mutations in sporadic primary melanomas: an inverse association with allelic loss on chromosome 9. Oncogene 2003;22:9217–24. [DOI] [PubMed] [Google Scholar]
- 23.Omholt K, Platz A, Kanter L, et al. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res 2003;9:6483–8. [PubMed] [Google Scholar]
- 24.Reifenberger J, Knobbe CB, Sterzinger AA, et al. Frequent alterations of Ras signaling pathway genes in sporadic malignant melanomas. Int J Cancer 2004;109:377–84. [DOI] [PubMed] [Google Scholar]
- 25.Shinozaki M, Fujimoto A, Morton DL, et al. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res 2004;10:1753–7. [DOI] [PubMed] [Google Scholar]
- 26.Uribe P, Wistuba II, Gonzalez S. BRAF mutation: a frequent event in benign, atypical, and malignant melanocytic lesions of the skin. Am J Dermatopathol 2003;25:365–70. [DOI] [PubMed] [Google Scholar]
- 27.Yazdi AS, Palmedo G, Flaig MJ, et al. Mutations of the BRAF gene in benign and malignant melanocytic lesions. J Invest Dermatol 2003;121:1160–2. [DOI] [PubMed] [Google Scholar]
- 28.Saldanha G, Purnell D, Fletcher A, et al. High BRAF mutation frequency does not characterize all melanocytic tumor types. Int J Cancer 2004;111:705–10. [DOI] [PubMed] [Google Scholar]
- 29.Yuen ST, Davies H, Chan TL, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res 2002;62:6451–5. [PubMed] [Google Scholar]
- 30.Jiveskog S, Ragnarsson-Olding B, Platz A, et al. N-ras mutations are common in melanomas from sun-exposed skin of humans but rare in mucosal membranes or unexposed skin. J Invest Dermatol 1998;111:757–61. [DOI] [PubMed] [Google Scholar]
- 31.Wong G, Pawelek J. Melanocyte-stimulating hormone promotes activation of pre-existing tyrosinase molecules in Cloudman S91 melanoma cells. Nature 1975;255:644–6. [DOI] [PubMed] [Google Scholar]
- 32.Halaban R, Pomerantz SH, Marshall S, et al. Tyrosinase activity and abundance in Cloudman melanoma cells. Arch Biochem Biophys 1984;230:383–7. [DOI] [PubMed] [Google Scholar]
- 33.Hunt G, Todd C, Cresswell JE, et al. Alpha-melanocyte stimulating hormone and its analogue Nle4DPhe7 alpha-MSH affect morphology, tyrosinase activity and melanogenesis in cultured human melanocytes. J Cell Sci 1994;107:205–11. [DOI] [PubMed] [Google Scholar]
- 34.Englaro W, Rezzonico R, Durand-Clement M, et al. Mitogen-activated protein kinase pathway and AP-1 are activated during cAMP-induced melanogenesis in B-16 melanoma cells. J Biol Chem 1995;270:24315–20. [DOI] [PubMed] [Google Scholar]
- 35.Englaro W, Bertolotto C, Busca R, et al. Inhibition of the mitogen-activated protein kinase pathway triggers B16 melanoma cell differentiation. J Biol Chem 1998;273:9966–70. [DOI] [PubMed] [Google Scholar]
- 36.Busca R, Abbe P, Mantoux F, et al. Ras mediates the cAMP-dependent activation of extracellular signal-regulated kinases (ERKs) in melanocytes. EMBO J 2000;19:2900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown AD, Lopez-Terrada D, Denny C, et al. Promoters containing ATF-binding sites are de-regulated in cells that express the EWS/ATF1 oncogene. Oncogene 1995;10:1749–56. [PubMed] [Google Scholar]
- 38.Fujimura Y, Ohno T, Siddique H, et al. The EWS-ATF-1 gene involved in malignant melanoma of soft parts with t(12;22) chromosome translocation, encodes a constitutive transcriptional activator. Oncogene 1996;12:159–67. [PubMed] [Google Scholar]
- 39.van Elsas A, Zerp SF, van der FS, et al. Relevance of ultraviolet-induced N-ras oncogene point mutations in development of primary human cutaneous melanoma. Am J Pathol 1996;149:883–93.8780392 [Google Scholar]


