Abstract
Four macrocyclic cystine-knot peptides of 29–31 residues, kalata, circulin A and B (CirA and CirB), and cyclopsychotride, have been isolated from coffee plants but have undetermined physiological functions. These macrocycles and 10 of their analogs prepared by chemical synthesis were tested against nine strains of microbes. Kalata and CirA were specific for the Gram-positive Staphylococcus aureus with a minimum inhibition concentration of ≈0.2 μM. They were relatively ineffective against Gram-negative bacteria such as Escherichia coli and Pseudomonas aeruginosa. However, CirB and cyclopsychotride were active against both Gram-positive and Gram-negative bacteria. In particular, CirB showed potent activity against E. coli with a minimum inhibitory concentration of 0.41 μM. All four cyclic peptides were moderately active against two strains of fungi, Candida kefyr and Candida tropicalis, but were inactive against Candida albicans. These macrocycles are cytotoxic and lysed human red blood cell with a lethal dose 50% of 400 μM. Modifying the Arg residue in kalata with a keto aldehyde significantly reduced its activity against S. aureus whereas blocking the arg in CirA produced no significant effect. The two-disulfide variants and their scrambled disulfide isomers exhibited antimicrobial profiles and potency similar to their native peptides. However, in high-salt assays (100 mM NaCl), few of these macrocyclic peptides, natives or analogs, retained antimicrobial activity. These results show that the macrocyclic peptides possess specific and potent antimicrobial activity that is salt-dependent and that their initial interactions with the microbial surfaces may be electrostatic, an effect commonly found in defensin antimicrobial peptides. Furthermore, their end-to-end cyclic structure with a cystine-knot motif represents a molecular structure of antimicrobials and may provide a useful template for the design of novel peptide antibiotics.
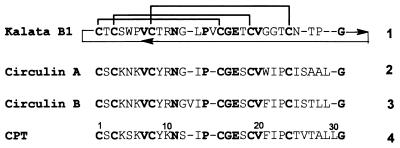
Although cyclic peptides are produced naturally in microbes and plants, they are usually restricted to small and medium size peptides of <15 amino acids. Thus, it is interesting to note the recent discovery of four large end-to-end cyclic peptides of 29–31 amino acids from plants of the Rubiacease family. These peptides contain six cysteines that can be aligned and share ≈45% of sequence homology (Fig. 1). As a group, they are reported to be the largest cyclic peptides found in nature. These include circulin A and B (CirA and CirB) from the African tropical tree Chassalia parvifolia (1), cyclopsychotride (Cpt) from the South American tropical plant Psychotria longipes (2), and kalata from the African plant Oldenlandia affinis (3, 4).
Figure 1.
Amino acid sequences of kalata, CirA, CirB, and Cpt. The conserved residues are indicated in bold. Sequence numbers are arbitrarily based on the linear precursors for chemical syntheses.
In addition to their cyclic nature, these macrocyclic peptides also have in common an inhibitor type of cystine-knot motif in their disulfide connectivity (4–8). This motif contains a bonding pattern of Cys I-IV, II-V, and III-VI. It is characterized by the Cys III-VI disulfide bond threading through the other two to give a knotted motif. The knotted disulfide motif together with small triple-stranded β-sheets have been found in peptide toxins and protease inhibitors of diverse origins (9). The constraints contributed by the cystine-knot disulfides and cyclic amide backbone found in the macrocyclic peptides confer resistance to enzymatic proteolysis, but this also complicates the elucidation of their disulfide connectivity by traditional methods. The initial evidence tentatively suggesting the cystine-knot arrangement of these peptides was found in kalata by using NMR methods and distance calculation (4). Subsequently, the cystine-knot motif of circulins was determined by partial acid hydrolysis and mass spectroscopy (5). Recently, our laboratory has confirmed the cystine-knot motifs of CirB and Cpt through total chemical synthesis and comparison with native samples obtained from plant extracts (6, 7).
CirA and CirB were identified through the anti-HIV drug screening program and were found to possess anti-viral activity (1). Both kalata and Cpt also were discovered through drug screening programs. Kalata shows uterotonic activity (3), and Cpt inhibits neurotensin binding to its receptor (2). Thus far, the physiological function of these macrocyclic peptides in plants remains unknown. However, comparisons with other peptides based on size, cystine-knot motif, or structural fold suggest that they may play a role in plant defense mechanisms (10–15).
The structures of kalata and CirA have been determined by two-dimensional NMR (4, 8). Both peptides are compact and fairly rigid structures exhibiting a similar global fold of distorted triple stranded β-sheets. The rigidity is contributed by the peptide backbone that is extensively folded back on itself and by the cross bracing of the three-disulfide bonds. The cystine-knot occupies most of the interior spaces and forms a sulfur-rich core. Computer modeling of kalata and CirA structures show that they display clusters of cationic charges and hydrophobic amino acids on their surfaces, characteristics of antimicrobial peptides (16, 17), and that has led us to hypothesize that these macrocyclic peptides may exhibit antimicrobial activity. To confirm this hypothesis, we have chemically prepared these four macrocyclic peptides and 10 two-disulfide variants. Our results confirm that they possess selective antimicrobial activity.
MATERIALS AND METHODS
Analytical HPLC was run on a Shimadzu system with a C18 Vydac column (250 × 4.6 mm) at a flow rate of 1 ml/min with a linear gradient of 0–85% buffer B (60% acetonitrile in H2O/0.04% trifluoroacetic acid; buffer A, 5% acetonitrile in H2O/0.045% trifluoroacetic acid) for 30 min with UV detection at 225 nm. Matrix-assisted laser desorption ionization/MS was performed on a Kompact MALD III instrument (Kratos Analytical Instruments). Samples were dissolved in 2 μl of a 1:1 mixture of H2O-CH3CN containing α-cyano-4-hydroxycinnamic acid. Measurements were made in the linear mode. All organisms were obtained from the American Type Culture Collection (ATCC). Gram-negative bacteria included Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Klebsiella oxytoca ATCC 49131, and Proteus vulgaris ATCC 49132. Two Gram-positive bacteria were Staphylococcus aureus 29213 and Micrococcus luteus ATCC 49732. Fungi included Candida albicans ATCC 37092, Candida kefyr ATCC 37095, and Candida tropicalis ATCC 37097. All strains were used for experiments for <4 weeks after taking from stock. The strains were incubated in trypticase soy broth (TSB), which was prepared in double distilled water and autoclaved for sterilization. TSB was purchased from Becton Dickinson. Liquid testing medium contained 10 mM sodium phosphate buffer (pH 7.4) and 3% TSB (wt/vol).
Peptide Synthesis.
Solid-phase peptide syntheses (18, 19) were performed manually or on an Applied Biosystems 430 synthesizer. The α-amines of all amino acids were protected with a Boc group.† The side-chain protections were as follows: Arg(Tos), Asn(Xan), Cys(4-MeBzl), Cys(Acm), Glu(OBzl), Lys(ClZ), Ser(Bzl), and Thr(Bzl). One cycle of the synthesis used 15–20 ml of solvent per gram of resin. Each cycle involved (i) a 20-min deprotection with 50% trifluoroacetic acid/CH2Cl2 and (ii) coupling with 4 equivalent each of Boc-amino acid and benzotrizol-1-yl-oxytris(dimethylamino)phosphonium hexafluorophosphate (20) in the presence of 12 equivalent N,N-diisopropylethylamine in dimethylformamide for 45 min. A typical procedure illustrated in the synthesis of CirA 2 was as follows (Fig. 2). CirA was synthesized on Boc-Gly-SCH2CH2CO–4-methyl benzhydrylamine resin 5 (940 mg, 0.26 mmol/g) (21, 22). Acetamidomethyl (Acm) was used to protected cysteine residue 3 and 19, and MeBzl was used for the other four residues. The thioester peptide was cleaved from the resin (250 mg) by high hydrofluoric acid (HF) (HF/anisole, 9:1, vol/vol) (23) to give the amide 6. The resulting peptides, still containing the two Cys(Acm) residues and the C terminus thioester, after the HF removal, were washed with diethylether to remove the organic scavengers. The unprotected and reduced peptide thioester NH2-CH(CH2SH)-CO-peptide-SCH2CH2CONH2 6 [molecular weight (MW) calculated (calc) 3404.7, found M+H+ 3406.5] was extracted into 8 M urea (150 ml) in phosphate buffer at pH 7.5 containing tris(carboxyethyl)phosphine (100 mg) to prevent polymeric disulfide formation. Sequential dialysis, first against 8 M urea (2000 ml) and then in three steps (in descending concentrations of 8 to 2 M urea solution) resulted in orthogonal cyclization. The whole process, monitored by analytical C18 reversed-phase HPLC and MS, was completed within 20 h. A single major peak of cyclic peptide 7 with four reduced thiols and two Cys(Acm) residues was found (MW calc 3299.7, found M+H+ 3300.5) in RP-HPLC. The dialyzed solution then was diluted to 1 M urea by water for the first-step oxidative folding by adding 15% DMSO (24, 25) to the peptide urea solution in 10 h to form a mixture of three two-disulfide intermediates 9a–c (MW calc 3295.7, found M+H+ 3296.8), which were separated by HPLC and whose disulfide connectivity was established by partial acid hydrolysis (26, 7). The third disulfide bond was formed by adjusting the pH to 4, removing the Acm groups, and oxidation. The solution was readjusted to pH 4 and bubbled with nitrogen for 10 min, followed by dropwise addition of iodine/methanol until a brown color persisted. The reaction, maintained in nitrogen atmosphere and in a darkened vessel, was completed in 45 min, as monitored by HPLC. The solution was cooled in an ice bath, and excess iodine was quenched by ascorbic acid. The desired cyclic peptide 2, purified by preparative RP-HPLC and characterized by MS giving the expected molecular weight (MW calc 3153.7, found M+H+ 3154.2), was obtained in 12% overall yield from the first resin bond amino acid. A similar procedure was used for synthesis of kalata 1 in 14% yield. MS gave the unprotected two-Acm peptide peptide thioester (MW calc 3145.3; found M+H+ 3147.1), cyclic peptide with four reduced thiols and two Acm (MW calc 3040.3; found M+H+ 3040.1), and kalata (MW calc 2892.3; found M+H+ 2893.2). Syntheses of CirB and Cpt were previously reported (6, 7).
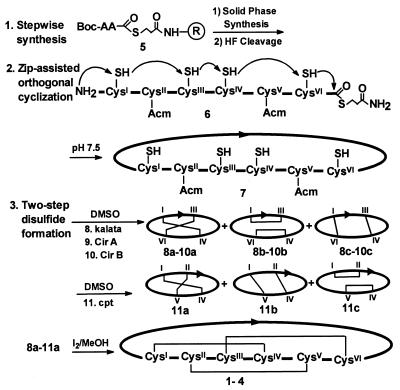
Figure 2.
Synthetic scheme for preparing the cystine-knot macrocycles and their analogs. Linear precursors were prepared by stepwise solid-phase synthesis. The freed peptides 6 without side chain protecting groups were cyclized in aqueous solutions at pH 7.5 through the thia zip reaction to afford macrocycles 7. The SS-protection schemes for kalata, CirA, and CirB were Cys II-V (Acm) to give the two-SS isomers 8–10 and Cys III-VI for Cpt to yield 11 after oxidation in 15% DMSO in aqueous buffer. Oxidation by I2 resulted in macrocycles 1–4.
Characterization.
Coelutions of CirA, CirB, and Cpt with the authentic samples isolated from natural sources (gifts from K. R. Gufatson, National Cancer Institute, and K. M. Witherup, Merck Pharmaceutical) were indistinguishable under several analytical HPLC systems. The disulfide connectivity was determined with the two-disulfide isomers 8–10 (0.12 mg) by partial acid hydrolysis in oxalic acid (0.25 M, 0.2 ml) at 100°C for 5 h. The hydrolysate was fractionated by HPLC, and the fragments were identified by matrix-assisted laser desorption ionization/MS.
Modifications of Arg in Kalata and CirA.
Purified synthetic kalata (2 mg) was modified with p-hydroxyphenylglyoxal (2 equivalent) in Tris⋅HCl buffer (pH 8.5) for 2 h in the dark at 20°C (27) to afford 12 in 70% yield (MW, calc 3042.2, M+H+ 3044.3, found). Arg10(Mts) CirA 13 (MW, calc 3478.1, M+H+ 3479.2, found) was obtained through total synthesis according to Fig. 2 by using low HF cleavage (23).
Antimicrobial Assays.
A sensitive and reproducible two-stage radial diffusion assay of Lehrer et al. (16) was used for testing the antimicrobial activity of cyclic peptides. A 1–4 × 106 colony-forming unit/ml of the organism was mixed with 10 ml of molten underlay gel solution and was poured into 10- × 10-cm Petri dishes (Nunc) to form a uniform layer. The gel solution contained 10 mM sodium phosphate buffer, 0.03% TSB, and 0.02% Tween 20. One gel solution had high salt (100 mM NaCl) whereas the other did not. After the gel solidified, gel wells were made by a 3-mm diameter template in an evenly spaced array. An aliquot of 5 μl of a serial half-log dilution of testing peptides at seven concentrations was added to each well after removing the gel plugs. The dishes were incubated at 37°C for 3 h to allow the peptides to diffuse into the underlay gels. Gels were overlaid with 10 ml of 1% agarose in 6% of TSB (wt/vol). After further incubation at 37°C for 16–24 h, the diameter of the clear zone surrounding the wells (colony-free) was measured under the microscope. Antimicrobial activities were expressed in units (0.1 mm = 1 unit), and the MICs were determined from the x intercepts of the dose-response curves.
Hemolysis and Cytotoxicity Assays.
Hemolytic activity of peptides was determined by using blood type A human erythrocytes (1.8 × 108 cells/ml). Peptide concentrations causing 50% hemolysis (EC50) were derived from the dose-response curves. Inhibition proliferation of mouse R1 fibroblasts by peptides was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (28) using 2–3 × 103 cells in a 96-well format for 2–3 days. Peptide concentrations causing 50% cell growth inhibition (IC50) were derived from the dose-response curves.
Computer Modeling.
Molecular modeling was done on a Silicon Graphics (Mountain View, CA) Irix 6.5 R10000 Workstation running charmm 22. Models were built by using MacKerell protein potential parameter sets. An initial conformation was constructed by assigning each residue the same backbone angles as the corresponding residue in the Kalata B1 pdb file (13). Minimization then was done by using 800 steps each (0.02-Å step size between updates) of the Steepest Descent and Conjugate Gradient algorithms adiabatically using a constant dielectric constant of 80 to approximate the presence of water. The final structure was visualized by using rasmol 2.6 for display (33).
RESULTS AND DISCUSSION
Synthetic Strategy.
Syntheses of all four cyclic peptides 1–4 were similar to those procedures previously developed by our laboratory to access end-to-end macrocyclic peptides (6, 7). They were achieved by a combination of three methods (Fig. 2): (i) a stepwise solid-phase synthesis (18) to obtain a linear peptide thioester 5 → 6; (ii) an end-to-end circularization 6→7 by an orthogonal cyclization method (22); and (iii) a two-step oxidation 7 → 1–4 to form the cystine-knot motif (24). Circularization of large peptides by the orthogonal cyclization method is more efficient and convenient than by conventional methods that usually require an elaborate scheme of protecting groups. It is performed in aqueous solutions by using unprotected peptide precursors without protection nor activation steps. When there are multiple Cys present in the linear precursor peptide sequence, such as those found in kalata and CirA, the cyclization process is assisted by a Thia Zip mechanism (7, 29) involving a series of thiol-thiolactone exchanges via side chain thiols 6. Eventually, an N-terminal (NT)-amino thiolactone intermediate is formed that links the NT-thiol with the C-terminal (CT)-carboxyl moiety as a large thiolactone. A spontaneous ring contraction of the NT-thiolactone through an S,N-acyl migration forms the end-to-end cyclic peptide. This reaction is highly regioselective and provides the necessary orthogonality to form the end-to-end amide bond between the NT-amine and the CT-thioester exclusively, even with unprotected precursors containing one or more of Lys or Glu, such as those found in CirA, CirB, and Cpt. The cyclic peptide then can be used directly, without purification, in the two-step disulfide oxidation method (24) for forming the cystine-knot motif.
Syntheses of Kalata and CirA.
The circular permutated nature of a cyclic peptide permits, in theory, the synthesis of a linear precursor at any given point. In practice, the choice of a linear precursor peptide sequence of these cyclic peptides in stepwise solid-phase synthesis is somewhat limited by the orthogonal cyclization method. This method requires a thiol of an NT-cysteine to displace the CT-thioester to give the N to CT-thiolactone intermediate (29). There are six cysteines in kalata and CirA. Although cyclization can be mediated through any one of these, the least hindered sites for cyclization between Cys-1 and Gly-29 in kalata (or between Cys-1 and Gly-30 in CirA) were chosen as the respective N-and C-terminal residues (Fig. 1). The peptide was prepared by solid-phase synthesis using Boc chemistry (18) on a thioester resin 5 containing a removable thiol alkyl linker (21, 22). The crude thioester peptide freed of its protecting groups was subjected to orthogonal cyclization in an aqueous solution buffered at pH 7.5. The cyclization of kalata or CirA was complete within 14 h and was quite rapid, considering that the cyclization involved a ring of >87 ring atoms. Detailed kinetic study in CirB showed that the circularization was assisted by a thia zip reaction (29) involving a series of thiol-thiolactone rearrangements in a cysteine-rich peptide to achieve end-to-end cyclization. The progress of the cyclization was monitored by HPLC and mass spectral analysis, and the whole process was complete within 15–20 h. The yield of the end-to-end cyclic peptides was ≈70%.
Disulfide bond formation of a cystine-knot motif posed a concern in the synthesis of CirA and kalata because an end-to-end cyclic peptide places all cysteines in close proximity. Indeed, in a random one-step oxidation, <5% of the desired correctly disulfide-paired product was obtained. To resolve this problem, we used a two-step disulfide-forming strategy (24) with a two-tiered protecting group scheme of 4-methylbenzyl (MeBzl) and acetamidomethyl (Acm). In the first step, the benzyl-based protecting group on the side-chain functional group, including the S-MeBzl, was cleaved by HF acidolysis during the release of the peptide from the resin support to afford 6. The Acm-thiol protecting group remained stable because of its resistance to acid. After the orthogonal cyclization, the Acm-protected pair in the cyclic peptide 7 then was liberated by a method that would not interfere with the stability of the previously formed disulfide bonds. An electrophillic agent such as I2 under acidic conditions was used to prevent disulfide rearrangement. Thus, the two-step method simplifies the synthetic scheme and reduces the possible disulfide isomers from 15 to 3. In the syntheses of 1–4, a nearly equal mixture of all three SS-isomers 8–11 was obtained in the first step. These two-disulfide intermediates were purified before oxidation by I2, and partial acid hydrolysis was used to determine their disulfide connectivity (7, 26). Furthermore, they also were used for structure-activity study. The overall yield of purified kalata and CirA was 12–14%. Purified kalata and CirA gave the expected molecular masses by matrix-assisted laser desorption ionization/MS analyses. Similar to CirB and Cpt (6, 7), synthetic CirA coeluted with the native CirA in HPLC whereas the other misformed isomers had different retention time and did not coelute with the authentic sample from the plant extract.
Antimicrobial Activity.
The MICs of these synthetic peptides against Gram-negative and Gram-positive bacteria and fungi were determined. Table 1 summarizes their antimicrobial activity in both high- and low-salt assays. In low-salt assays, kalata showed a strong antimicrobial activity against Gram-positive bacteria, particularly against S. aureus with a MIC of 0.26 μM. It was moderately active against Gram-negative bacterium K. oxytoca and fungus C. kefyr.
Table 1.
Antimicrobial activity of kalata, CirA, Cir B, and Cpt in high- and low-salt assays
| Organism | MIC, μM
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Kalata
|
CirA
|
CirB
|
Cpt
|
|||||
| H-salt | L-salt | H-salt | L-salt | H-salt | L salt | H-salt | L salt | |
| E. coli | >500 | >500 | >500 | >500 | >500 | 0.41 | >500 | 1.55 |
| Pse. aeruginosa | >500 | >500 | >500 | >500 | 48.0 | 25.5 | 50.2 | 13.5 |
| Pr. vulgaris | >500 | >500 | >500 | 54.6 | >500 | 6.80 | >500 | 13.2 |
| K. oxytoca | >500 | 54.8 | >500 | >500 | 15.6 | 8.20 | 13.2 | 5.80 |
| S. aureus | >500 | 0.26 | >500 | 0.19 | >500 | 13.5 | >500 | 39.0 |
| M. luteus | >500 | 40.4 | >500 | >500 | >500 | >500 | >500 | 48.0 |
| C. albicans | >500 | >500 | >500 | >500 | >500 | >500 | >500 | >500 |
| C. kefyr | >500 | 21.4 | >500 | 18.6 | >500 | 29.0 | 48.0 | 14.0 |
| C. tropicalis | >500 | >500 | >500 | 19.4 | >500 | >500 | >500 | 56.5 |
Experiment were performed in radial diffusion assay with underlay gel containing 1% agarose and 10 mM phosphate buffer with (high-salt) or without (low-salt) 100 mM NaCl. H, high; L, low.
The antimicrobial profile of CirA was comparable to kalata. Both were active against four microbes being tested. CirA was potent against S. aureus with a MIC at 0.19 μM, although it was inactive against M. luteus. Like kalata, it exhibited little activity against Gram-negative bacteria. It was moderately active against C. kefyr and C. tropicalis with MICs at ≈19 μM.
Although the structure and net positive charges of CirB are very similar to CirA, their antimicrobial profiles are significantly different. CirB is broader in antimicrobial spectrum than CirA and was active against six of the nine microbes tested in our assays bacteria. CirB exhibited strong activity against all four Gram-negative bacteria tested in this study. It was especially potent against E. coli with a MIC at 0.41 μM. In contrast, it was only moderately active against the Gram-positive bacterium S. aureus.
Among the four macrocyclic peptide tested, Cpt showed the broadest spectrum of antimicrobial activity. It was active in eight of the nine microbes being tested, including both Gram-positive and -negative bacteria and two fungi. In terms of potency, Cpt was comparable to CirB against E. coli and other Gram-negative bacteria. Similar to the other three macrocyclic peptides, it was not active against C. albicans.
Most of the antimicrobial activity was abrogated when tested in high-salt conditions, with two exceptions. Activity of CirB or Cpt was largely unaffected by the salt concentrations against two Gram-negative bacteria, Pse. aeruginosa and K. oxytoca, and Cpt retained its activity against C. kefyr. The potency of kalata, CirA, CirB, and Cpt against some specific microbes, such as S. aureus and E. coli, at low μM level is comparable to those other cystine-rich and β-stranded antimicrobial peptides, such as defensins (16).
Hemolytic Activity and Cytotoxicity.
Concentrations of all macrocycles 1–4 causing 50% red blood cell hemolysis were >400 μM. Their activities roughly correlated with their antimicrobial potency, with CirB and Cpt being more hemolytic (550 and 405 μM, respectively) than kalata and CirA (1510 and 1020 μM, respectively). CirB and Cpt were also cytotoxic, causing 50% cell growth inhibition of mouse fibroblasts at 820 and 1850 μM, respectively.
Structure-Activity Study.
To test the requirement of the disulfide bonds, the two-disulfide intermediates of kalata, Cir B, and Cpt obtained during the course of our syntheses were tested for their antimicrobial activity. In kalata 8a, or CirB 10a, the disulfide bond of Cys II-V was deleted, and the two Cys were acetamidomethylated while the Cys III-VI pair was similarly deleted in Cpt 11a. Table 2 shows the effect of deleting a disulfide on the antimicrobial activity of all four cyclic peptides. Overall, the antimicrobial profiles were unaltered by the deletion of one disulfide bond in these rigid cyclic peptides. However, potency decreases of 2- to 3-fold were observed in 25% of these analogs.
Table 2.
Antimicrobial activity of two-disulfide analogs in low-salt conditions
| Organism | MIC, μM
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Kalata
|
CirA
|
CirB
|
Cpt
|
||||||
| 8a | 12 | 13 | 10a | 10b | 10c | 11a | 11b | 11c | |
| E. coli | >500 | >500 | >500 | 0.33 | 0.45 | 0.57 | 2.80 | 1.55 | 0.98 |
| Pse. aeruginosa | >500 | >500 | >500 | 48.0 | 50.2 | 50.2 | 17.5 | 48.0 | 13.5 |
| Pr. vulgaris | 48.5 | NT | 45.5 | 14.8 | 48.0 | 18.2 | 15.6 | 18.4 | 14.8 |
| K. oxytoca | 44.6 | 56.5 | NT | 10.5 | 20.4 | 18.6 | 5.60 | 5.20 | 17.4 |
| S. aureus | 0.29 | 33.5 | 0.21 | 48.0 | 13.5 | 29.5 | 7.90 | 48.0 | 13.5 |
| M. luteus | 14.2 | >500 | >500 | >500 | >500 | >500 | 8.0 | 48.0 | 13.2 |
| C. albicans | >500 | >500 | >500 | >500 | >500 | >500 | >500 | >500 | >500 |
| C. kefyr | 19.2 | 48.0 | 51.4 | 29.0 | 18.5 | 48.0 | 48.0 | 106 | 29.0 |
| C. tropicalis | >500 | >500 | 49.8 | >500 | >500 | >500 | >500 | >500 | >500 |
All peptides also were tested in high-salt conditions (100 mM NaCl) and were found to be inactive at MIC >500 μM except CirB 10c, Cpt 11c, and Cpt 11b against K. oxytoca and C. kefyr with MIC of 29.0–56.5 μM. NT, not tested.
Three geometric disulfide isomers of CirB 10 and Cpt 11 were obtained during the oxidative folding by the two-step oxidation method (Fig. 2). Table 2 summarizes the effect of scrambled disulfide bonds of CirB 10b, c and Cpt 11b, c on the antimicrobial activity under low-and high- salt conditions. Comparing with the native molecules 3 and 4 (Table 1) or the correctly paired two-disulfide variants 10a and 11a, the scrambling of disulfide bonds in these isomers does not result in any change of antimicrobial profile or significant loss of activity. It appears that the end-to-end macrocyclic motif preorganizes the structures and that deleting a SS-bond or scrambling of the cystine-knot disulfides does not significantly alter their antimicrobial activity. These results are consistent with our recent study on α-defensins, in which an end-to-end macrocyclic structure can be used to replace the native three-disulfide structure without significant loss of antimicrobial activity.
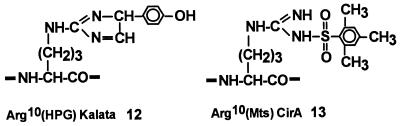
The importance of cationic charges was tested in analogs 12, 13 of kalata and CirA (Table 2). We used two methods to modify the arginine residue in these two cyclic peptides. In the first method, purified synthetic kalata 1 was chemoselectively modified by an Arg-specific reagent. Blocking the sole cationic Arg10 of kalata with a keto aldehyde, p-hydroxyphenylglyoxal (27), resulted in a kalata analog 12 with a modified guanidino group at Arg10 (Fig. 3). This modification sharply reduced the antimicrobial activity against M. luteus and C. kefyr and >100 fold against S. aureus. The Arg10 in CirA was modified by a second method. During solid phase synthesis, all side-chain protecting groups were protected, and these protecting groups were removed concurrently from the resin support by a high HF procedure (23). However, the mesitylenesulfonyl (Mts) protecting group on Arg is known to be more acid stable than other benzyl alcohol-derived protecting groups. As a result, by manipulating a less acidic deprotection condition, such as low HF (23), a linear peptide thioester with an Arg10(Mts) (but without other side-chain protecting groups) was obtained. Orthogonal cyclization as shown in Fig. 2 afforded a two-disulfide Arg10(Mts) analog 13 (Fig. 3). Reducing the cationic charge of Arg10 in CirA by this electron-withdrawing, Mts-protecting group did not result in any significant loss of activity against either E. coli or Pr. vulgaris. However, there is ≈2.5-fold loss of activity against C. kefyr and C. tropicalis. Because the antimicrobial profiles of kalata with a lone Arg and CirA with three cationic amino acids are quite similar, these results suggest that at least a cationic residue is required for electrostatic interaction with bacterial surfaces for activity. Further analogs are needed to confirm this conclusion.
Figure 3.
Modification of Arg10 of kalata by p-hydroxyphenyl glyoxal at pH 8.5 to afford Arg10(Hpg) kalata and Arg10 of CirA by mesitylenesulfonylation during synthesis that resulted in Arg10(Mts) CirA.
Computer Modeling of Electrostatic Interaction by Cationic Charges.
Computer models of CirB and Cpt are based on the reported structures of kalata (4) and CirA (8). Fig. 4 shows the backbone structure, the cystine-knot, the charge, and hydrophobic distribution of these four cyclic peptides. All four compounds show a distorted spherical globular shape. The interior is filled by three disulfide bonds of the cystine knot that contributes to the compactness of these molecules. The distributions of charge and hydrophobic residues on the surfaces of these cyclic peptides are clustered. The cationic charge(s) are located along one edge of the molecules whereas the hydrophobic residues that are solvent-exposed are clustered on one face.
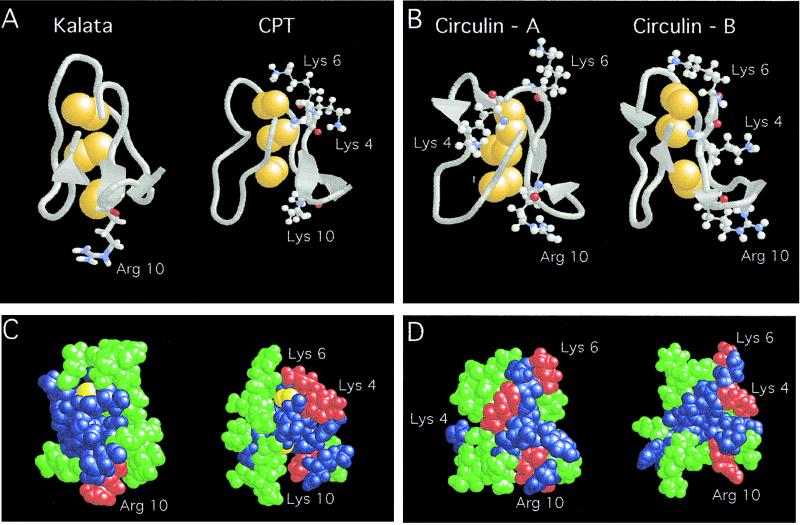
Figure 4.
Computer modeling of kalata, CirA, CirB, and Cpt. (A and B) The backbone structures with cationic amino acids (Arg and Lys) in ball-and-stick figures. Arg10 located at the bottom is used as a reference point. The cystine-knot is shown in yellow. (C and D) Space-filling models showing clusters of hydrophobic in green, hydrophilic in blue, and cationic amino acids in red.
Because most cationic peptide antibiotics exhibit the characteristic charge and hydrophobic residues clustering pattern, we attempted to use this model to interpret our results on the specificity of these cyclic peptides and their analogs against Gram-positive (kalata and CirA-specific) and Gram-negative bacteria (CirB and Cpt-specific). A simple explanation is the electrostatic model of interaction (17). Gram-negative bacteria contain an outer membrane rich in lipid A, which are full of ionic charges. Polycationic charges clustering is required in peptide antibiotics, such as defensins, cercopins, maganins, and lipopolysaccharide-binding proteins (16, 17, 30, 31), to be effective in interaction with Gram-negative bacteria. CirA, CirB, and Cpt are cationic in nature. Each contains a net of two positive charges with three cationic amino acids within the sequence of KXKVCYR10/K (Fig. 1). However, the cationic nature of kalata is not apparent because it contains only a cationic Arg10. Thus, it is not surprising to find CirB and Cpt containing a cluster of three cationic amino acids to be active against Gram-negative bacterium whereas kalata containing a lone cationic Arg10 is inactive. Modifying this cationic charge significantly diminishes its activity of kalata against Gram-positive bacteria whereas removing one of the three cationic charges of CirA is far less deleterious. Similar to the disulfide-rich family of defensins (16), the antimicrobial activity is salt-dependent, an effect that is also consistent with the electrostatic interaction model. Although both CirA and CirB contain a cluster of three cationic amino acids, the 29-aa CirA is inactive against the Gram-negative bacteria, E. coli. A plausible explanation may lie in the orientations of these cationic charges. Arg10 in CirA is folded back to its backbone and is shielded from Trp20, which is absent in CirB. Furthermore, the 30-aa CirB contains an additional amino acid in its sequence, Val13. Such an insertion appears to change the conformation of the cationic clusters so that they are more exposed and aligned continuously on one edge of the molecule. Such a flexible cluster containing three-cationic amino acids also is observed in Cpt, which may explain their activity against Gram-negative bacteria.
CONCLUSIONS
Over the past 20 years, >150 peptide antibiotics from nonmicrobial sources have been discovered (16, 17). These antimicrobial peptides derived from the higher forms of organisms (plants, insects, and animals) are usually cationic, amphipathic, and in open chain forms (secondary structure). Our results show that the four recently discovered macrocyclic peptides with similar sequences and sizes from coffee plants of the Rubiacease family can be included as antimicrobial peptides. All four macrocycles tested in nine strains of microbes show significant activity against Gram-positive and negative bacteria, moderate activity against two fungi strains, and no activity against C. albicans. Kalata and CirA are particularly potent against S. aureus, whereas CirB and Cpt are active against both Gram-positive and Gram-negative bacteria. We also show that, despite their cyclic structures and cystine-knot disulfide motif, they are amenable to chemical synthesis through the orthogonal cyclization method. Furthermore, chemical synthesis provides a useful approach to confirm their structures. These macrocyclic peptides are different from the known, open-chained plant defensins that are larger in size. Thus, they represent a class of structural motifs for antimicrobial peptides with rigid, compact, and macrocyclic structures. These structural features may be useful for understanding the antimicrobial mechanisms of peptide antibiotics and may lead to design of peptide mimetics.
Acknowledgments
This work was in part supported by U.S. Public Health Service Grants CA36544 and GM 57145.
ABBREVIATIONS
- Acm
acetamidomethyl
- ATCC
American Type Culture Collection
- CirA
circulin A
- Cpt
cyclopsychotride
- CT
C-terminal
- MIC
minimum inhibitory concentration
- Mts
mesitylene-2-sulfonyl
- MW
molecular weight
- NT
N-terminal
- TSB
tryptic soy booth
Footnotes
Throughout the paper, standard abbreviations are used for the amino acids and protecting groups (32).
References
- 1.Gustafson K R, Sowder R C, II, Henderson L E, Parsons I C, Kashman Y, Cardellina J H, II, McMahon J B, Buckheit R W, Jr, Pannell L K, Boyd M R. J Am Chem Soc. 1994;116:9337–9338. [Google Scholar]
- 2.Witherup K M, Bogusky M J, Anderson P S, Ramjit H, Ransom R W, Wood T, Sardana M. J Nat Prod. 1994;57:1619–1625. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- 3.Gran L. Acta Pharmacol Toxicol. 1973;32:400–408. doi: 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 4.Saether O, Craik D J, Campbell I D, Sletten K, Juul J, Norman D G. Biochemistry. 1995;34:4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 5.Derua R, Gustafson K R, Pannell L K. Biochem Biophys Res Commun. 1996;228:632–638. doi: 10.1006/bbrc.1996.1708. [DOI] [PubMed] [Google Scholar]
- 6.Tam J P, Lu Y-A. Tetrahedron Lett. 1997;38:5599–5602. [Google Scholar]
- 7.Tam J P, Lu Y-A. Protein Sci. 1998;7:1583–1592. doi: 10.1002/pro.5560070712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly N L, Koltay A, Gustafson K R, Boyd M R, Casas-Finet J R, Craik D J. J Mol Biol. 1999;285:333–345. doi: 10.1006/jmbi.1998.2276. [DOI] [PubMed] [Google Scholar]
- 9.Pallaghy P K, Niellsen K J, Craik D J, Norton R S. Protein Sci. 1994;3:1833–1839. doi: 10.1002/pro.5560031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holak T A, Gondol D, Otlewski J, Wilusz T. J Mol Biol. 1989;210:635–648. doi: 10.1016/0022-2836(89)90137-x. [DOI] [PubMed] [Google Scholar]
- 11.Bindoli A, Rigobello M P, Favel A. J Neurochem. 1988;50:138–141. doi: 10.1111/j.1471-4159.1988.tb13240.x. [DOI] [PubMed] [Google Scholar]
- 12.Atkinson A H, Heath R L, Simpson R J, Clarke A E, Anderson M A. Plant Cell. 1993;5:203–213. doi: 10.1105/tpc.5.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen K J, Hearth R L, Anderson M A, Craik D J. Biochemistry. 1995;34:14304–14311. doi: 10.1021/bi00044a007. [DOI] [PubMed] [Google Scholar]
- 14.Vervoort J, van den Hooven H W, Berg A, Vossen P, Vogelsang R, Joosten M H, de Wit P J. FEBS Lett. 1997;404:153–158. doi: 10.1016/s0014-5793(97)00117-8. [DOI] [PubMed] [Google Scholar]
- 15.Broekaert W F, Terras F R, Cammue B P, Osborn R W. Plant Physiol. 1995;108:1353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehrer R I, Rosenman M, Harwing S S, Jackson R, Eisenhauer P. J Immunol Methods. 1991;137:167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 17.Hancock R E W, Falla T, Brown M. Adv Microb Physiol. 1995;37:135–175. doi: 10.1016/s0065-2911(08)60145-9. [DOI] [PubMed] [Google Scholar]
- 18.Merrifield R B. Science. 1986;57:1619–1625. [Google Scholar]
- 19.Wade D, Boman A, Wahlin B, Drain C M, Andreu D, Boman H G, Merrifield R B. Proc Natl Acad Sci USA. 1990;87:4761–4765. doi: 10.1073/pnas.87.12.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro B, Dormoy J R, Evin G, Selve C. Tetrahedron Lett. 1975;14:1219–1222. [Google Scholar]
- 21.Hojo H, Aimoto S. Bull Chem Soc Jpn. 1991;64:111–117. [Google Scholar]
- 22.Zhang L, Tam J P. J Am Chem Soc. 1997;119:2363–2370. [Google Scholar]
- 23.Tam J P, Heath W F, Merrifield R B. J Am Chem Soc. 1983;105:6442–6455. [Google Scholar]
- 24.Yang Y, Sweeney W V, Schneider K, Chait B, Tam J P. Protein Sci. 1994;3:1267–1275. doi: 10.1002/pro.5560030813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam J P, Wu C R, Liu W, Zhang J W. J Am Chem Soc. 1991;113:6657–6622. [Google Scholar]
- 26.Zhou Z, Smith D L. J Protein Chem. 1990;9:523–532. doi: 10.1007/BF01025005. [DOI] [PubMed] [Google Scholar]
- 27.Yamasaki R B, Vega A, Feeney R E. Anal Biochem. 1980;109:32–40. doi: 10.1016/0003-2697(80)90006-8. [DOI] [PubMed] [Google Scholar]
- 28.Carmichael J, DeGraff W G, Gazdar A F, Minna J D, Mitchell J B. Cancer Res. 1987;47:936–942. [PubMed] [Google Scholar]
- 29.Tam J P, Lu Y-A, Yu Q. J Am Chem Soc. 1999;121:4316–4324. [Google Scholar]
- 30.Elsbach P, Weiss J, Kao L. J Biol Chem. 1985;260:1618–1122. [PubMed] [Google Scholar]
- 31.Beamer L J, Carroll S F, Eisenberg D. Protein Sci. 1998;7:906–914. doi: 10.1002/pro.5560070408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.IUPAC-IUB Commission for Biochemical Nomenclature. J Biol Chem. 1985;260:14–42. [Google Scholar]
- 33.Sayle R. rasmol 2.6.Molecular Visualization Program. Stevenage, U.K.: Glaxo Wellcome Research and Development; 1995. [Google Scholar]