Abstract
Background: Individual differences in T cell responsiveness to interleukin 12 (IL-12), resulting from inherited factors, may be responsible for differences in the intensity of cell mediated immune (CMI) responses in patients with leprosy, a disease with a wide clinical spectrum.
Aim: Polymorphisms in the 5′ flanking region of the IL12RB2 gene were analysed to determine potential immunogenetic factors affecting CMI responses, using leprosy as a model.
Methods: Polymorphisms in the 5′ flanking region of IL12RB2 were examined using direct sequencing techniques, and allele frequencies between patients with lepromatous leprosy and patients with tuberculoid leprosy were compared. The effect of these single nucleotide polymorphisms (SNPs) on IL12RB2 expression was estimated using the dual luciferase reporter gene assay in Jurkat T cells.
Results: Several SNPs, including −1035A>G, −1023A>G, −650delG, and −465A>G, were detected within the 5′ flanking region of IL12RB2. The frequency of haplotype 1 (−1035A, −1023A, −650G, −464A) was high in the general Japanese population, but was significantly lower in lepromatous patients compared with tuberculoid patients and healthy controls. Reporter gene assays using Jurkat T cells revealed that all haplotypes carrying one or more SNP exhibited a lower transcriptional activity compared with haplotype 1.
Conclusion: SNPs within the 5′ flanking region of IL12RB2 affect the degree of expression of this gene and may be implicated in individual differences in CMI responsiveness to mycobacterial antigens, leading to lepromatous or tuberculoid leprosy.
Keywords: single nucleotide polymorphisms, IL12RB2, cell mediated immune responses, leprosy, mycobacterial antigens
The importance of the cell mediated immune (CMI) response is well established in the host defence to mycobacterial pathogens.1,2 Leprosy, a chronic disease caused by infection with Mycobacterium leprae, shows a wide spectrum of clinical features.3 Patients with tuberculoid type leprosy (T-lep) show a high CMI response to M leprae, with resistance to infection, whereas patients with lepromatous leprosy (L-lep) show a poor CMI response to the pathogen and have a progressive form of the disease. Although a leishmaniasis model using BALB/c and C57BL6 mice has improved our understanding of cellular and genetic control mechanisms for infectious diseases and allergy, no such models have yet been established in humans. We propose leprosy as an alternative model in humans.
“Polymorphisms in the 5′ flanking region of IL12RB2 may affect the expression of the interleukin 12 receptor β2 chain, resulting in individual differences in the intensity of cell mediated immune responses to mycobacteria”
Interleukin 12 (IL-12) is secreted from macrophages and dendritic cells and is a potent inducer of interferon γ production by T helper type 1 (Th1) cells, which is in part dependent upon the degree of expression of the IL-12 receptor (IL-12R) on the cell surface.4–6 IL-12R is composed of two protein subunits, referred to as the β1 and β2 chains, and expression of the β2 chain is a crucial determinant of Th1/Th2 balance, because STAT4 is activated through interaction with a tyrosine residue on the cytoplasmic domain of the IL-12R β2 subunit.7,8,9,10 It has been shown that the expression of IL-12Rβ2 is greater in tuberculoid lesions than in lepromatous lesions, whereas the expression of IL-12Rβ1 is similar in both.11 We hypothesise that the susceptibility to several diseases related to mycobacterial pathogens could be determined by the degree of expression of IL-12Rβ2, which might be regulated by genetic factors, including IL12RB2 polymorphism.
One hypothesis is that polymorphisms in the 5′ flanking region of IL12RB2 may affect the expression of IL-12Rβ2, resulting in individual differences in the intensity of CMI responses to mycobacteria. We examined single nucleotide polymorphisms (SNPs) within the 5′ flanking region of IL12RB2 as feasible markers to determine susceptibility to the disease and the effect of these SNPs on the transcription of IL-12Rβ2 molecules.
MATERIALS AND METHODS
Study population
Genomic DNA samples were collected from 176 Japanese patients with leprosy—130 with L-lep and 46 with T-lep—and 68 healthy Japanese donors. Patients were clinically diagnosed, according to the description of Ridley and Jopling, referring to results of the Mitsuda test and their sequelae.3 Donors were recruited into our study under informed consent guidelines approved by the human ethical committee of Saitama Medical School, Japan.
Analysis of polymorphism in the 5′ flanking region of IL12RB2
Genomic DNA was sequenced for polymorphisms in the 5′ flanking region of IL12RB2 using direct sequencing. Briefly, a fragment spanning −1247 to +55 of IL12RB2 was polymerase chain reaction (PCR) amplified and subsequently sequenced using the ABI 3730 DNA sequencer (PerkinElmer Life Sciences, Wellesley, Massachusetts, USA). The sequence data obtained were compared with the GenBank database (GenBank accession number AL389925) to determine the SNPs on IL12RB2. Numbers of base positions were defined as the distance from the start point of the reported cDNA sequence.12 The haplotypes were determined by cloning the PCR products into the pGEM-T Easy plasmid (Promega, Madison, Wisconsin, USA) and sequencing.
Evaluation of transcriptional activity of the 5′ flanking region of IL12RB2
For transfection studies, the dual luciferase reporter gene assay system (Promega) was used with the IL12RB2–pGL3 plasmid constructs. The plasmids comprised a 1.3 kb NheI/HindIII digested PCR fragment of IL12RB2 ligated to the pGL3 basic vector (Promega). Jurkat T cells (1 × 107) were electroporated with 25 μg of plasmid DNA (IL12RB2–pGL3 constructs) and a control Renilla luciferase reporter plasmid (pRL–TK, 25 ng; Promega) using a Gene Pulser (Bio-Rad Laboratories, Hercules, California, USA), as described previously.13 Cells were then cultured for 48 hours in the presence of antihuman CD3 monoclonal antibody (PharMingen, San Diego, California, USA), and antihuman CD28 monoclonal antibody (PharMingen) for the final 24 hour period. Cell lysates were prepared, and both firefly and Renilla luciferase activities were evaluated using a dual luminometer Fluoroskan Ascent FL (Thermo Electron Oy, Vantaa, Finland).
RESULTS
Analysis of polymorphism in the 5′ flanking region of IL12RB2
Twelve SNPs located within the 5′ flanking region of IL12RB2 (−1247 to +55), comprising −1047delT, −1035A>G, −1033T>C, −1023A>G, −650delG, −568A>C, −557T>C, −550T>C, −464A>G, −464A>C, −202T>C, and −188A>C, were identified. The surveillance study of 176 patients (130 with L-lep and 46 with T-lep) revealed significant differences in frequencies of SNPs between these patients groups; in particular: −1035A>G, −1023A>G, −650delG, and −464A>G (table 1).
Table 1.
Allelic distribution in patients with leprosy
| SNPs | Clinical form | Allele frequency | OR | RR | p Value |
| −1035A>G | L-lep | 24.6% | 3.97 | 3.24 | <0.001 |
| T-lep | 7.6% | ||||
| −1023A>G | L-lep | 24.2% | 2.95 | 2.48 | <0.01 |
| T-lep | 9.8% | ||||
| −650delG | L-lep | 28.8% | 3.74 | 2.95 | <0.001 |
| T-lep | 9.8% | ||||
| −464A>G | L-lep | 23.1% | 3.64 | 3.03 | <0.01 |
| T-lep | 7.6% |
Frequencies of four single nucleotide polymorphisms (SNPs) were compared between lepromatous (L-lep) and tuberculoid (T-lep) patients. The χ2 test was used to compare differences in the distribution of clinical phenotypes and allele frequencies. The odds ratio (OR) and relative risk (RR) were also calculated by comparing the frequency of variant alleles between two clinical types of leprosy.
We randomly selected 17 healthy donors heterozygous in at least one of the positions −1035, −1023, −650, and −464 to determine linkage disequilibrium. Five haplotypes were determined by sequencing 34 subcloned PCR amplified DNA fragments from the 17 heterozygous donors. Haplotype 1 consisted of −1035A, −1023A, −650G, and −464A, and haplotype 2 consisted of −1035G, −1023G, −650del, and −464G, and accounted for 73.6% of the haplotypes detected in our study (table 2).
Table 2.
Haplotypes of the 5′ flanking region of IL12RB2
| Haplotype | −1035 | −1023 | −650 | −464 | Frequency (%) |
| 1 | A | A | G | A | 41.2 |
| 2 | G | G | del | G | 32.4 |
| 3 | A | A | del | A | 11.8 |
| 4 | G | G | G | A | 8.3 |
| 5 | A | A | del | G | 5.9 |
Haplotypes were determined by sequencing 34 subcloned polymerase chain reaction amplified DNA fragments from 17 donors with heterozygous alleles in at least in one of the positions −1035, −1023, −650, and −464, but not from donors carrying homozygous alleles at all positions.
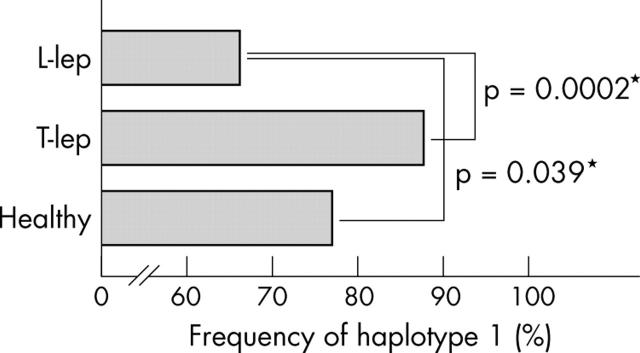
Haplotype frequency was calculated based on the assumption that each group would be in accordance with the Hardy–Weinberg equilibrium because subjects were selected from the Japanese Wajin population of mainland Japan (Honshu). p Values were calculated using the StatView (SAS Institute, Cary, North Carolina, USA) statistical software program, by comparing the frequency of haplotype 1 in patients with L-lep leprosy, those with T-lep leprosy, and healthy controls. The frequency of haplotype 1 was significantly lower in patients with L-lep compared with those with T-lep and healthy donors (fig 1), suggesting that haplotype 1 might contribute to the intensity of CMI responses to mycobacteria.
Figure 1.
Frequency (%) of haplotype 1 in the three groups: patients with lepromatous leprosy (L-lep), patients with tuberculoid type leprosy (T-lep), and healthy controls. The haplotype frequency was calculated based on the assumption that each group would be in accordance with the Hardy–Weinberg equilibrium because subjects were selected from the Japanese Wajin population of mainland Japan (Honshu). p Values were calculated using the StatView statistical software program, by comparing the frequency of haplotype 1 in the L-lep leprosy, T-lep leprosy, and healthy control groups.
Evaluation of transcriptional activity of the 5′ flanking region of IL12RB2
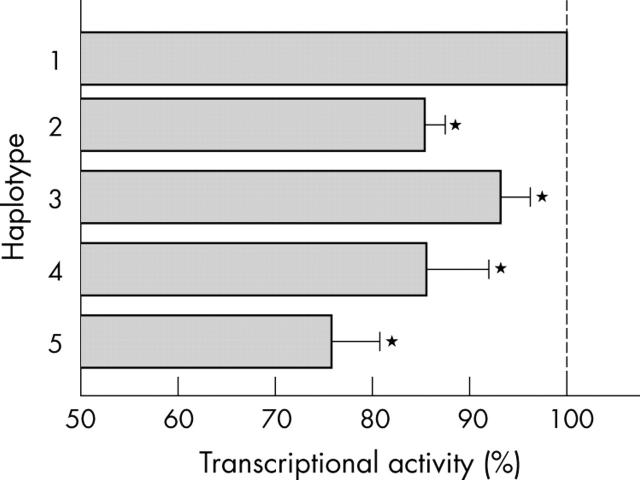
The transcriptional activity of each reporter construct was determined using the dual luciferase reporter gene assay. The transcriptional activity of haplotype 1 was significantly higher than haplotypes 2, 3, 4, and 5 (fig 2). Each experiment was performed using triplicate wells and was repeated four times. Because each SNP is a genetic factor potentially able to reduce the expression of IL-12Rβ2 molecules, a poor CMI response to mycobacteria may occur.
Figure 2.
Basal promoter activity of reporter constructs containing −1247 to +55 of IL12RB2 harbouring each haplotype. The transcriptional activity of each haplotype is indicated by the percentage of relative luciferase units compared with the haplotype 1 construct. Each experiment was assayed using triplicate wells and was repeated four times and similar results were obtained. *p < 0.05; χ2 test.
DISCUSSION
It was recently reported that the lack of IL-12Rβ1 expression caused by mutations in IL12RB1 resulted in human immunodeficiency, thereby demonstrating the essential role of IL-12 in resistance to infections caused by intracellular bacteria.14–16 However, no differences in the expression of IL-12Rβ1 on T cells were detected between donors, including patients and healthy subjects (data not shown). It has been reported that IL-12Rβ2 is absent in freshly isolated peripheral blood mononuclear cells, whereas up to 72% of resting peripheral blood mononuclear cells from normal volunteers express IL-12Rβ1 molecules, and that IL-12Rβ2 is expressed selectively in Th1 cells but not in Th2 cells.9,17 Moreover, because IL-12Rβ2 has tyrosine residues in the cytoplasmic domain that play a role in signal transduction, we hypothesised that IL-12Rβ2, but not IL-12Rβ1, could be important in explaining the low CMI responses induced by IL-12 in T cells from patients with leprosy.
“Individual differences in the intensity of the cell mediated immune response to mycobacteria are probably regulated primarily by the degree of expression of IL-12Rβ2, rather than possible conformational changes”
Alternatively spliced mRNA with the absence of IL12RB2 exon 15 leads to the loss of induction of interferon γ production.18 Similarly, we have previously found several coding SNPs of IL12RB2, but could not determine an effect on CMI response intensity or on alternatively spliced mRNA (data not shown). Epidemiological studies recently performed by two independent groups demonstrated that there was no influence of IL12RB2 coding SNPs on susceptibility to mycobacterial infection.19,20 Therefore, individual differences in the intensity of the CMI response to mycobacteria are probably regulated primarily by the degree of expression of IL-12Rβ2, rather than possible conformational changes caused by the coding SNPs detected in the Japanese population. Bleharski et al showed differences in gene expression profiles according to the clinical type of leprosy,21 and the IL-12R gene was included among those differentially expressed between patients with L-lep and T-lep.
The differences in transcriptional activity between haplotype 1 and other haplotypes are marginal, except for that between haplotypes 1 and 5. These findings suggest that haplotype 5 might be more closely associated with susceptibility to lepromatous leprosy than haplotypes 2, 3, and 4. To investigate this issue, it will be necessary to analyse the frequency of all haplotypes in the patient population. Moreover, the functional effects of SNPs −650 and −464 on the transcriptional mechanism should also be elucidated.
Take home messages.
Single nucleotide polymorphisms (SNPs) within the 5′ flanking region of the IL12RB2 gene could affect the expression of the interleukin 12 receptor β2 chain (IL-12Rβ2) and result in the individual differences in the intensity of cell mediated immune responses that lead to the lepromatous and tuberculoid types of leprosy
Haplotype 5 appears to be more closely associated with susceptibility to lepromatous leprosy than haplotypes 2, 3, and 4, but further investigations are necessary
These SNPs may also affect susceptibility to allergy because IL-12Rβ2 is involved in the allergic response
An SNP at position −464 of IL12RB2 was found to have high transcriptional activity compared with the wild-type allele, possibly because of disruption of a GATA site.22 This appears to contradict our present data, although the effects of SNPs on the binding affinity with GATA-3 were not directly tested in this report. It was determined that the main target of GATA-3 is not IL-12Rβ2, but rather STAT4.23 Further studies are needed to determine the precise molecular mechanism.
Taken together, we conclude that SNPs within the 5′ flanking region of IL12RB2 could affect the expression of IL-12Rβ2, thus causing individual differences in the intensity of CMI responses leading to the lepromatous and tuberculoid types of leprosy. It is probable that these SNPs also affect susceptibility to allergy, because IL-12Rβ2 is implicated in the Th1/Th2 balance in the allergic response. A study investigating this hypothesis is currently under way.
Acknowledgments
This study was supported, in part, by a grant from the US–Japan Cooperative Medical Science Program Tuberculosis and Leprosy Panel, a Health Sciences Research Grant for Research on Emerging and Re-emerging Infectious Diseases, Nagao Memorial Fund, and a grant from the Ochiai Memorial Award 2003.
Abbreviations
CMI, cell mediated immunity
IL-12, interleukin 12
IL-12R, interleukin 12 receptor
L-lep, lepromatous leprosy
PCR, polymerase chain reaction
SNP, single nucleotide polymorphism
Th, T helper
T-lep, tuberculoid type leprosy
REFERENCES
- 1.Bloom BR, Godal T. Selective primary health care: strategies for control of disease in the developing world. V. Leprosy. Rev Infect Dis 1983;5:765–80. [DOI] [PubMed] [Google Scholar]
- 2.Sieling PA, Modlin RL. Cytokine patterns at the site of mycobacterial infection. Immunobiology 1994;191:378–87. [DOI] [PubMed] [Google Scholar]
- 3.Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis 1966;34:255–73. [PubMed] [Google Scholar]
- 4.Trinchieri G. Interleukin-12: a cytokine produced by antigen-presenting cells with immunoregulatory functions in the generation of T-helper cells type 1 and cytotoxic lymphocytes. Blood 1994;84:4008–27. [PubMed] [Google Scholar]
- 5.Sieling PA, Wang XH, Gately MK, et al. IL-12 regulates T helper type 1 cytokine responses in human infectious disease. J Immunol 1994;153:3639–47. [PubMed] [Google Scholar]
- 6.Gately MK, Renzetti LM, Magram J, et al. The interleukin-12/interleukin-12-receptor system: role in normal and pathologic immune responses. Annu Rev Immunol 1998;16:495–521. [DOI] [PubMed] [Google Scholar]
- 7.Rogge L, Barberis-Maino L, Biffi M, et al. Selective expression of an interleukin-12 receptor component by human T helper 1 cells. J Exp Med 1997;185:825–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabo SJ, Dighe AS, Gubler U, et al. Regulation of the interleukin (IL)-12R β2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J Exp Med 1997;185:817–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogge L, Papi A, Presky DH, et al. Antibodies to the IL-12 receptor β2 chain mark human Th1 but not Th2 cells in vitro and in vivo. J Immunol 1999;162:3926–32. [PubMed] [Google Scholar]
- 10.Naeger LK, McKinney J, Salvekar A, et al. Identification of a STAT4 binding site in the interleukin-12 receptor required for signaling. J Biol Chem 1999;274:1875–8. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Uyemura K, Van Dyke MK, et al. A role for IL-12 receptor expression and signal transduction in host defense in leprosy. J Immunol 2001;167:779–86. [DOI] [PubMed] [Google Scholar]
- 12.Presky DH, Yang H, Minetti LJ, et al. A functional interleukin 12 receptor complex is composed of two β-type cytokine receptor subunits. Proc Natl Acad Sci U S A 1996;93:14002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Rietschoten JG, Smits HH, van de Wetering D. et al. Silencer activity of NFATc2 in the interleukin-12 receptor β2 proximal promoter in human T helper cells. J Biol Chem 2001;276:34509–16. [DOI] [PubMed] [Google Scholar]
- 14.de Jong R, Altare F, Haagen IA, et al. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science 1998;280:1435–8. [DOI] [PubMed] [Google Scholar]
- 15.Sakai T, Matsuoka M, Aoki M, et al. Missense mutation of the interleukin-12 receptor beta1 chain-encoding gene is associated with impaired immunity to Mycobacterium avium complex infection. Blood 2001;97:2688–94. [DOI] [PubMed] [Google Scholar]
- 16.Altare F, Ensser A, Breiman A, et al. Interleukin-12 receptor beta1 deficiency in a patient with abdominal tuberculosis. J Infect Dis 2001;184:231–6. [DOI] [PubMed] [Google Scholar]
- 17.Wu CY, Warrier RR, Carvajal DM, et al. Biological function and distribution of human interleukin-12 receptor beta chain. Eur J Immunol 1996;26:345–50. [DOI] [PubMed] [Google Scholar]
- 18.van Rietschoten JG, Smits HH, Westland R, et al. Genomic organization of the human interleukin-12 receptor β2-chain gene. Immunogenetics 2000;51:30–6. [DOI] [PubMed] [Google Scholar]
- 19.Lee SB, Kim BC, Jin SH, et al. Missense mutations of the interleukin-12 receptor beta 1 (IL12RB1) and interferon-gamma receptor 1 (IFNGR1) genes are not associated with susceptibility to lepromatous leprosy in Korea. Immunogenetics 2003;55:177–81. [DOI] [PubMed] [Google Scholar]
- 20.Akahoshi M, Ishihara M, Remus N, et al. Association between IFNA genotype and the risk of sarcoidosis. Hum Genet 2004;114:503–9. [DOI] [PubMed] [Google Scholar]
- 21.Bleharski JR, Li H, Meinken C, et al. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science 2003;301:1527–30. [DOI] [PubMed] [Google Scholar]
- 22.van Rietschoten JG, Westland R, van den Bogaard R, et al. A novel polymorphic GATA site in the human IL-12Rβ2 promoter region affects transcriptional activity. Tissue Antigens 2004;63:538–46. [DOI] [PubMed] [Google Scholar]
- 23.Usui T, Nishikomori R, Kitani A, et al. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rβ2 chain or T-bet. Immunity 2003;18:415–28. [DOI] [PubMed] [Google Scholar]