Abstract
Background: Over 90% of Ewing’s sarcoma/primitive neuroectodermal tumour (ES/PNET) cases have the t(11;22) chromosomal rearrangement, which is also found in other small round cell tumours, including desmoplastic small round cell tumour (DSRCT) and clear cell sarcoma (CCS). Although this rearrangement can be analysed by fluorescence in situ hybridisation (FISH) using routinely formalin fixed, paraffin wax embedded (FFPE) tissues when fresh or frozen tissues are not available, a sensitive and convenient detection method is needed for routine clinical diagnosis.
Aims: To investigate the usefulness of newly developed probes for detecting EWS rearrangement resulting from chromosomal translocations using FISH and FFPE tissue in the clinical diagnosis of ES/PNET, DSRCT, and CCS.
Methods: Sixteen ES/PNETs, six DSRCTs, and six CCSs were studied. Three poorly differentiated synovial sarcomas, three alveolar rhabdomyosarcomas, and three neuroblastomas served as negative controls. Interphase FISH analysis was performed on FFPE tissue sections with a commercially available EWSR1 (22q12) dual colour, breakapart rearrangement probe.
Results: One fused signal and one split signal of orange and green, demonstrating rearrangement of the EWS gene, was detected in 14 of 16 ES/PNETs, all six DRSCTs, and five of six CCSs, but not in the negative controls.
Conclusions: Interphase FISH using this newly developed probe is sensitive and specific for detecting the EWS gene on FFPE tissues and is of value in the routine clinical diagnosis of ES/PNET, DSRCT, and CCS.
Keywords: Ewing’s sarcoma/primitive neuroectodermal tumour, desmoplastic small round cell tumour, clear cell sarcoma, fluorescence in situ hybridisation, formalin fixed paraffin wax embedded
Ewing’s sarcoma and primitive neuroectodermal tumour (ES/PNET) are mainly composed of small round cells and are highly malignant, usually affecting the bone and extraosseous tissues of children and young adults. Diagnosing ES/PNET accurately is sometimes arduous, particularly in patients with unusual clinical features, such as disease onset in old age and/or an organ based origin. The identification of the highly specific balanced chromosomal rearrangement t(11;22)(q24;q12) in most ES/PNETs1,2 provides a valuable tool for diagnosing this tumour at the molecular level. The application of fluorescence in situ hybridisation (FISH) methodology has resulted in a significant improvement in diagnostic ability with the establishment of specific DNA probes capable of detecting chromosomal aberrations in formalin fixed, paraffin wax embedded (FFPE) tissue in some soft tissue sarcomas.3,4,5,6,7,8,9,10,11 Recently, probes adjoining or spanning the Ewing’s sarcoma breakpoint region 1 (EWSR1) gene on 22q12 have been developed and used in FISH assays to detect this translocation, and provide a reliable, accurate, and relatively simple diagnostic approach on FFPE tissue.5,7,8,9,10,11 However, there is still the need for more sensitive and specific probes, and the relatively costly and time consuming process needs to be adapted for routine clinical diagnosis.
A commercially available EWSR1 (22q12) dual colour, breakapart rearrangement probe was developed recently and has been tested extensively by Vysis (http://www.vysis.com) before release. Several studies have reported the application of this probe, which has proved to have practical advantages over earlier probes.12–14
“Diagnosing Ewing’s sarcoma/primitive neuroectodermal tumour accurately is sometimes arduous, particularly in patients with unusual clinical features, such as disease onset in old age and/or an organ based origin”
Here, we report our findings using this newly developed probe for the detection of the EWS rearrangement on interphase nuclei extracted from FFPE tissue, and we demonstrate the usefulness of using this technique in the clinical diagnosis of ES/PNET and other small round cell tumours sharing the EWS rearrangement, including desmoplastic small round cell tumour (DSRCT) and clear cell sarcoma (CCS).
MATERIALS AND METHODS
Specimen selection
Archival tumour specimens were retrieved from the pathological files at the laboratory of pathology, National Cancer Centre (NCC), Tokyo, Japan. A series of small round cell tumours with morphological and immunophenotypical resemblance diagnosed and treated at the centre was selected for study. Tumours included 16 primary ES/PNETs (10 seen in house and six in consultation), six DSRCTs, and six CCSs. Three alveolar rhabdomyosarcomas, three poorly differentiated synovial sarcomas, and three neuroblastomas served as negative controls because there are no EWS rearrangements in these tumour types. For light microscopic study, all specimens were routinely fixed in 10% formalin and embedded in paraffin wax, and 4 μm thick sections were then cut and stained with haematoxylin and eosin. Frozen tissues were available for reverse transcribed polymerase chain reaction (RT-PCR) analysis in seven of 16 ES/PNETs, five of six CCSs, and in all cases of DSRCT, alveolar rhabdomyosarcoma, and poorly differentiated synovial sarcoma.
We defined ES/PNET as a small round cell tumour; focal rosette-like arrangement of tumour cells was seen in some lesions (fig 1A). All tumours were consistently immunopositive for CD99 (O-13). Of 16 ES/PNETs, frozen tissue was available from seven at the time of diagnosis for RT-PCR analysis, and the EWS–FLI1 fusion gene was detected in five. There were five male and 11 female patients, with ages at the time of diagnosis ranging from 11 to 72 years (mean, 39). The sites of involvement were the bone in four patients, kidney in two, duodenum and pancreas in one patient each, and other soft tissue with various anatomical locations in eight patients, two of which occurred in subcutaneous locations (cases 4 and 14) (table 1).
Figure 1.
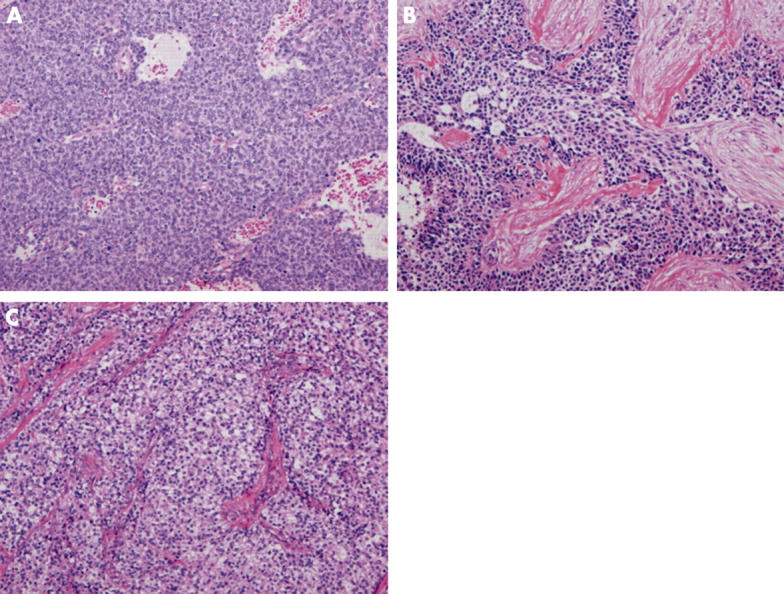
(A) Low power view of a Ewing’s sarcoma and primitive neuroectodermal tumour of the pancreas, composed of a monotonous proliferation of small round cells, with an undifferentiated appearance (haematoxylin and eosin; original magnification, ×100). (B) Low power view of an intra-abdominal desmoplastic small round cell tumour. Compact nests consisting of small cells separated by abundant desmoplastic stroma, with an undifferentiated appearance, can be seen (haematoxylin and eosin; original magnification, ×100). (C) A clear cell sarcoma showing nested epithelioid cells with clear cytoplasm and prominent nucleoli. The nests of tumour cells were separated by fibrous bands (haematoxylin and eosin; original magnification, ×100).
Table 1.
Clinical findings of the 16 ES/PNETs
| Case | Age (years) | Sex | Tumour site | Tumour size (cm) | RT-PCR | FISH | Follow up (months) | Outcome |
| 1 | 11 | M | Femur | 10.0 | EWS–FLI1 | Positive | 32 | DOD |
| 2 | 15 | M | Thigh | 16.0 | ND | Positive | 11 | DOD |
| 3 | 18 | F | Rib | 4.0 | EWS–FLI1 | ND* | 56 | CDF |
| 4 | 19 | M | Thigh | 8.0 | EWS–FLI1 | Positive | 75 | CDF |
| 5 | 28 | M | Kidney | NA | NA | Positive | NA | NA |
| 6 | 28 | F | Humerus | 9.0 | ND | ND* | 6 | CDF |
| 7 | 31 | F | Retroperitoneum | 6.0 | NA | Positive | 10 | DOD |
| 8 | 32 | F | Duodenum | 6.0 | NA | Positive | 24 | DOD |
| 9 | 37 | F | Pancreas | NA | NA | Positive | NA | NA |
| 10 | 43 | F | Sacrum | 7.5 | NA | Positive | 3 | AWD |
| 11 | 52 | M | Epidura | NA | EWS–FLI1 | Positive | NA | NA |
| 12 | 56 | F | Retroperitoneum | NA | NA | Positive | NA | NA |
| 13 | 57 | F | Chest wall | 6.0 | NA | Positive | 4 | AWD |
| 14 | 60 | F | Thigh | 4.1 | EWS–FLI1 | Positive | 8 | CDF |
| 15 | 61 | F | Kidney | NA | NA | Positive | NA | NA |
| 16 | 72 | F | Retroperitoneum | 5.0 | NA | Positive | 3 | AWD |
*Decalcified tissue was used.
AWD, alive with disease; CDF, continued disease free; DOD, died of disease; ES/PNET, Ewing’s sarcoma and primitive neuroectodermal tumour; FISH, fluorescence in situ hybridisation; NA, data not available; ND, not detected; RT-PCR, reverse transcription polymerase chain reaction.
The DSRCTs were classified primarily on the basis of compact nests consisting of small round cells separated by abundant desmoplastic stroma (fig 1B), with an undifferentiated appearance, and had strong immunoreactivity for vimentin, broad spectrum keratin (AE1/3), epithelial membrane antigen, desmin, and neurone specific enolase. In all cases, desmin had a distinctive, intense, intracytoplasmic location that was often dot-like. The diagnosis was further confirmed by the presence of the EWS–WT1 fusion gene demonstrated by RT-PCR analysis. Table 2 summarises the clinical findings of the six DSRCTs.
Table 2.
Clinical findings of the 6 intra-abdominal desmoplastic small round cell tumours
| Case | Age (years) | Sex | Tumour site | RT-PCR | FISH | Follow up (month) | Outcome |
| 1 | 22 | M | Omentum and mesentery | EWS–WT1 | Positive | 6 | DOD |
| 2 | 18 | M | Omentum and mesentery | EWS–WT1 | Positive | 15 | DOD |
| 3 | 20 | M | Omentum and mesentery | EWS–WT1 | Positive | 27 | DOD |
| 4 | 32 | M | Omentum and mesentery | EWS–WT1 | Positive | 28 | DOD |
| 5 | 35 | M | Omentum and mesentery | EWS–WT1 | Positive | 12 | AWD |
| 6 | 28 | M | Omentum and mesentery | EWS–WT1 | Positive | 10 | AWD |
AWD, alive with disease; DOD, died of disease; FISH, fluorescence in situ hybridisation; RT-PCR, reverse transcription polymerase chain reaction.
The CCSs were composed of compact nests or fascicles of rounded or fusiform cells with clear cytoplasm (fig 1C). All of these tumours were immunohistochemically positive for HMB-45 and S-100 protein and five of six cases were cytogenetically confirmed by detecting EWS–ATF1 fusion transcripts (table 3).
Table 3.
Clinical findings of the 6 clear cell sarcomas analysed
| Case | Age (years) | Sex | Tumour site | Tumour size (cm) | RT-PCR | FISH | Follow up (months) | Outcome |
| 1 | 25 | M | Thigh | 4.0 | EWS–ATF1 | Positive | 24 | AWD |
| 2 | 33 | M | Kidney | 12.0 | EWS–ATF1 | Positive | 8 | DOD |
| 3 | 38 | M | Abdominal wall | 6.0 | NA | Positive | 252 | DOD |
| 4 | 41 | M | Forearm | 4.5 | EWS–ATF1 | Positive | 6 | AWD |
| 5 | 62 | F | Ankle | 4.0 | EWS–ATF1 | ND | 20 | CDF |
| 6 | 69 | F | Hand | NA | EWS–ATF1 | Positive | 49 | DOD |
AWD, alive with disease; CDF, continued disease free; DOD, died of disease; FISH, fluorescence in situ hybridisation; NA, data not available; ND, not detected; RT-PCR, reverse-transcription polymerase chain reaction.
The rhabdomyosarcomas and poorly differentiated synovial sarcomas were defined morphologically and immunohistochemically, and confirmed by the presence of the PAX3/PAX7–FKHR and SYT–SSX fusion genes by RT-PCR analysis, respectively.
RT-PCR analysis
For the molecular genetic analysis, total RNA was extracted from the frozen tumour tissues using standard procedures.15 The RNA was reverse transcribed, and the samples were then subjected to PCR amplification using a previously described pair of primers for EWS and FLI1,16 EWS–WT1,17 EWS–ATF1,18 PAX3/PAX7 consensus and FKHR,19 and SSX and SYT.20 The PCR was performed according to the methodology described previously in each tumour type.16–20
Fluorescence in situ hybridisation
FISH was carried out according to the manufacturer’s instructions. Briefly, after dewaxing the slides, they were immersed in 0.2N HCl for 20 minutes and pretreatment solution (Vysis, Downer’s Grove, Illinois, USA) at 80°C for 30 minutes, digested with protease for 60 minutes at 37°C, washed in 1× phosphate buffered saline for five minutes at room temperature, fixed in 10% formaldehyde for 10 minutes at room temperature, washed in 1× phosphate buffered saline for five minutes at room temperature, placed into prewarmed denaturation solution (Vysis) for five minutes at 72°C, and then dehydrated by immersing in 70%, 85%, and 100% ethanol for one minute each at room temperature.
For interphase FISH, the slides were subjected to hybridisation with an LSI EWSR1 (22q12) dual colour, breakapart rearrangement probe (Vysis). The probe consists of a mixture of two FISH DNA probes. The first probe is an ∼ 500 kb probe labelled with spectrum orange; it flanks the 5′ side of the EWSR1 gene (22q12) and extends inward into intron 4. The second one is an ∼ 1100 kb probe labelled with spectrum green; it flanks the 3′ side of the EWSR1 gene (22q12). The FISH probe mix (10 µl) was added to the sample area of the slides at 45°C. The slides were coverslipped, sealed with rubber cement, and incubated at 37°C for 48 hours in a humidified chamber. The slides were then washed in posthybridisation wash buffer at 72°C. Subsequently, 10 µl of DAPI counterstain was placed on the slide, which was then coverslipped. After hybridisation, all slides were maintained at −20°C in the dark. Hybridisation signals were visualised with an epifluorescence microscope, and images were captured on a CCD camera. Fifty nuclei that showed both green and orange signals were counted by two different individuals, and the percentages of green, orange, and fused signals were calculated.
RESULTS
Molecular genetic features
Frozen tissues were available from seven of 16 ES/PNETs, and EWS–FLI1 fusion transcripts were detected in five of these seven samples. PCR products revealed inframe fusions between EWS exon 7 and FLI1 exon 5 in four tumours (cases 1, 3, 11, and 14), and EWS exon 7 and FLI1 exon 6 in one tumour (case 4) (table 1). In two cases, the fusion transcripts were not detected (cases 2 and 6). In the six DSRCTs, the PCR products revealed inframe fusions of EWS exon 9 to WT1 exon 8 in three tumours (cases 1, 2, and 4), of EWS exon 10 to WT1 exon 8 in two tumours (cases 3 and 6), and of EWS exon 7 to WT1 exon 8 in the remaining tumour (case 5) (table 2). An amplified PCR product corresponding to the EWS–ATF1 fusion was detected in five CSSs, three of which were EWS exon 8 to ATF1 exon 4 (cases 1, 5, and 6) and two (cases 2 and 4) were EWS exon 10 to ATF1 exon 5 (table 3). A 583 bp amplification product corresponding to the SYT–SSX fusion gene and an 870 bp amplification product consistent with PAX3–FKHR gene fusion was detected in three poorly differentiated synovial sarcomas and three alveolar rhabdomyosarcomas, respectively.
FISH analysis
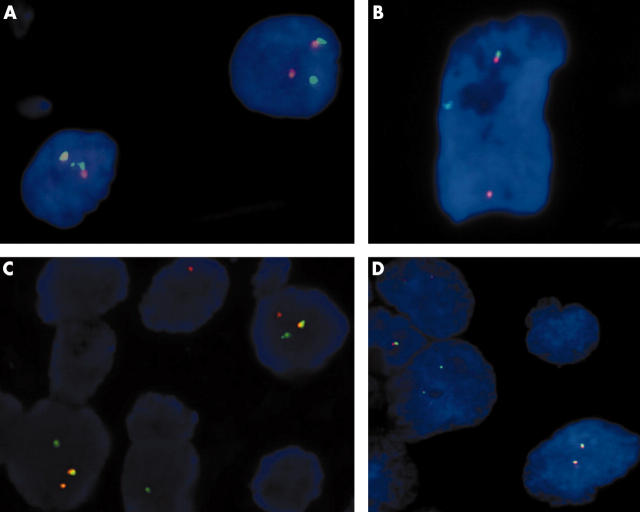
Fusion of orange and green signal patterns, as seen in negative control tumour cells, was indicative of an intact copy of the EWS gene. A separated signal pattern of one green and one orange, demonstrating a rearrangement of the EWS gene, was detected in 50–90% of nuclei in 14 of 16 ES/PNETs (table 1) (fig 2A). There was no rearrangement of the EWS gene found in two cases (cases 3 and 9) in which the tissues were decalcified. Fusion signals were detected in more than 90% of the nuclei in all six DRSCTs and in 45–90% of the nuclei in five of the six CCSs specimens (fig 2B, C), but in no cases of alveolar rhabdomyosarcoma, poorly differentiated synovial sarcoma, or neuroblastoma (fig 2D).
Figure 2.
Results of hybridisation of the EWSR1 dual colour breakapart probes. Tumour cells of (A) a Ewing’s sarcoma and primitive neuroectodermal tumour, (B) a desmoplastic small round cell tumour, and (C) a clear cell sarcoma show one fusion, one orange, and one green signal pattern, indicative of a rearrangement of one copy of the EWSR1 region. (D) Tumour cells of an alveolar rhabdomyosarcoma show two fusion signal patterns, reflecting the two intact copies of EWSR1.
DISCUSSION
RT-PCR analysis is a powerful tool for the diagnosis of tumours with chromosomal translocations. However, the limitation of RT-PCR analysis for routine diagnosis is that fresh or frozen tissue is sometimes not available, and that the fusion transcripts are not always detectable with RT-PCR using FFPE tissue. Moreover, it is necessary to prepare various primers, and care needs to be taken to avoid false positive results caused by contamination when carrying out these processes. As seen in our series and that of previous workers,21,22 ES/PNET can rarely occur in old age and in other anatomical locations, such as the kidney, duodenum, and pancreas. Diagnosing this tumour in such unusual locations is difficult, and it is necessary to separate it from other solid cancers and malignant lymphoma. Although immunohistochemical analysis of CD99 can provide a clue to the differential diagnosis in this situation, cytogenetic analysis is needed for a definite diagnosis because the expression of CD99 is not specific for ES/PNETs.23
Several investigators have used the FISH technique to identify the 11;22 translocation in ES/PNETs.5,6,7,8,9,10 However, most of these have been performed on cell lines or touch imprints and short term cultures, and sometimes centromeric and total probes need to be prepared or a computer based statistical analysis is required. These limit the routine clinical use of this technique, as discussed previously.11 In a previous study, a new set of probes specific to both the EWS and FLI1 breakpoint regions was reported to be useful in clinical diagnosis using free nuclear cytospins from FFPE tissue.11 However, this method is relatively labour intensive, and there is the need to eliminate random overlap of signals in non-ES/PNETs.
The newly developed probes that we used were localised to the breakpoints on chromosome 22q12 and provided evidence of the t(22q12) translocation by showing one orange and one green signal pattern on the derivative chromosome 22. The signals generated by these probes were generally large, bright, and easily detectable, and could be detected even when the fusion gene was not detected by RT-PCR (table 1, case 2). These probes provide a definite advantage in the routine clinical diagnosis of screening tumours that share rearrangement of the EWS gene. The average percentage of positive results using these probes for detecting rearrangement of the EWS gene on three tumour types was 90%, indicating an excellent performance. The probes performed extremely well—rearrangements of the EWS gene were detected in approximately 90% of the three tumour types tested. However, this suggests that up to 10% of tumours would not be detected if this test alone was used in clinical diagnosis. Fusion signals were not detected in two ES/PNETs, perhaps because only decalcified tissue was available as the tumours had not extended into the soft tissue in these cases (cases 3 and 6). One case of RT-PCR confirmed CCS was FISH negative; the reasons for the failure of FISH analysis in this case are unclear, but there may have been technical problems because FISH analysis is a multistep procedure.
The gene fusion EWS–FLI1, resulting from t(11;22)(q24;q12), is seen in approximately 85% of cases of ES/PNET. In our series, five of seven (71%) tumours had the EWS–FLI1 translocation. Variant fusions of EWS with other ETS family genes—ERG (at 21q22), ETV1 (7p22), E1AF (17q12), and FEV (2q33)—have been found in rare cases of ES/PNET.24 It is possible that in the two cases (cases 2 and 6) in which we could not detect EWS–FLI1 fusion transcripts, the EWS gene was fused with other ETS variants. Such rearrangements of the EWS gene in ES/PNET were detected in one of the two cases by these probes in FFPE tissue sections.
Although the probes we used specifically identify t(22q12) but cannot specifically identify the translocation partners, there are advantages to detecting t(22q12) translocation by using one probe only: using several probes is costly and time consuming. To distinguish between small round cell tumours sharing EWS rearrangements, it may be necessary to combine both histopathological and immunohistochemical approaches. Moreover, this FISH technique may give either false positive or false negative results, so that it should be carried out together with other cytogenetic analyses, including RT-PCR, in diagnostically challenging cases.
“The signals generated by these probes were generally large, bright, and easily detectable, and could be detected even when the fusion gene was not detected by reverse transcriptase polymerase chain reaction”
The intra-abdominal DSRCT with multiphenotypic differentiation is a recently identified but well characterised entity with distinctive clinical, light microscopic, and immunohistochemical features.25–30 Cytogenetic studies of DSRCTs have described a consistent chromosomal translocation, t(11;22)(p13;q12), which creates a novel rearrangement of the EWS genes at 22q12 and WT1 at 11p13.17,28–30 Variants of t(11;22) have been demonstrated in a small number of DSRCTs, including a t(2;21;22) translocation,31 an unusual case that had a single cell with a 22q deletion and a single cell with an 11p13 deletion,32 and a t(11;22;21) translocation that showed an EWS–WT1 fusion transcript.33 Fusion in these cases should also generate a target detectable by these probes. It is usually difficult to separate ES/PNET from DSRCT morphologically, although they do have several distinguishing features. Immunohistochemical markers such as keratin, neural antigens, and desmin would be useful in the differential diagnosis of these two tumour types.34
CCS is a rare and aggressive tumour, arising predominantly from the soft tissue of the extremities in young adults, and should be separated from ES/PNET. Most CCSs have a distinctive chromosomal rearrangement t(12;22)(q13;q12), associated with a EWS–ATF1 fusion.35–37 Unlike ES/PNETs, most cases of CCS have round to ovoid vesicular nuclei, with prominent basophilic nucleoli and clear or pale staining cytoplasm. Immunohistochemically, CCSs are CD99 negative, nearly all cases are S-100 positive, and most show positivity for melanocytic markers (tyrosinase, melan-A, and HMB-45).
Take home messages.
We devised a fluorescence in situ hybridisation (FISH) technique on formalin fixed, paraffin wax embedded (FFPE) samples of Ewing’s sarcoma/primitive neuroectodermal tumour (ES/PNET) and other small round cell tumours using a commercially available EWSR1 (22q12) dual colour, breakapart rearrangement probe
Interphase FISH using this newly developed probe is sensitive and specific for detecting the EWS gene on FFPE tissues and is useful in the routine clinical diagnosis of ES/PNET and other small round cell tumours, such as desmoplastic small round cell tumour and clear cell sarcoma
To distinguish between small round cell tumours sharing EWS rearrangements, it may be necessary to combine both histopathological and immunohistochemical approaches
Finally, it may be difficult to discriminate CCSs from malignant melanomas, although melanomas are generally more polymorphic and rarely show pale or clear cytoplasm. The results of immunohistochemistry are often confusing because the immunophenotypes of these two tumour types are very similar. In this situation, detecting an EWS gene rearrangement specific for CCS by FISH analysis, as described here, is a valuable tool for the differential diagnosis of the two tumour types.
In conclusion, the interphase FISH method using EWSR1 dual colour breakapart probes is sensitive and specific for the detection of rearrangement of the EWS gene on chromosome 22q12. This method is a useful aid in detecting chromosomal t(11;22) translocation variants in FFPE tissue and will be of value in the clinical diagnosis of ES/PNET and other small round cell tumours sharing EWS rearrangements, including DSRCT and CCS.
Acknowledgments
This work was supported by a Grant-in-Aid for Cancer Research (16-6) from the Ministry of Health, Labor, and Welfare of Japan. We thank K Yokozawa and M Gotoh for their technical assistance. We also thank M Ishiguro for her helpful comments on the FISH procedures.
Abbreviations
CCS, clear cell sarcoma
DSRCT, desmoplastic small round cell tumour
ES/PNET, Ewing’s sarcoma/primitive neuroectodermal tumour
EWSR1, Ewing’s sarcoma breakpoint region 1
FFPE, formalin fixed, paraffin wax embedded
FISH, fluorescence in situ hybridisation
PCR, polymerase chain reaction
RT, reverse transcribed
REFERENCES
- 1.Aurias A, Rimbaut C, Buffe D, et al. Translocation involving chromosome 22 in Ewing’s sarcoma. A cytogenetic study of four fresh tumors. Cancer Genet Cytogenet 1984;12:21–5. [DOI] [PubMed] [Google Scholar]
- 2.Turc-Carel C, Philip I, Berger MP, et al. Chromosomal study of Ewing’s sarcoma (ES) cell lines. Consistency of a reciprocal translocation t(11;22)(q24;q12). Cancer Genet Cytogenet 1984;12:1–19. [DOI] [PubMed] [Google Scholar]
- 3.Schofield DE, Fletcher JA. Trisomy 12 in pediatric granulosa-stromal cell tumors: demonstration by a modified method of fluorescence in situ hybridization on paraffin-embedded material. Am J Pathol 1992;141:1265–9. [PMC free article] [PubMed] [Google Scholar]
- 4.Lee W, Han K, Harris CP, et al. Use of FISH to detect chromosomal translocations and deletions: analysis of chromosomal rearrangement in synovial sarcoma cells from paraffin-embedded specimens. Am J Pathol 1993;143:15–19. [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor C, Patel K, Jones T, et al. Diagnosis of Ewing’s sarcoma and peripheral neuroectodermal tumour based on the detection of t(11;22) using fluorescence in situ hybridization. Br J Cancer 1993;67:128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagao K, Gomyo Y, Ito H, et al. A case report of synovial sarcoma with translocation t(X;18). Application of fluorescence in situ hybridization to paraffin-embedded tissue. Virchows Arch 1995;426:519–22. [DOI] [PubMed] [Google Scholar]
- 7.Desmaze C, Zucman J, Delattre O, et al. Unicolor and bicolor in situ hybridization in the diagnosis of peripheral neuroepithelioma and related tumors. Genes Chromosomes Cancer 1992;5:30–4. [DOI] [PubMed] [Google Scholar]
- 8.Desmaze C, Zucman J, Delattre O, et al. Interphase molecular cytogenetics of Ewing’s sarcoma and peripheral neuroepithelioma t(11;22) with flanking and overlapping cosmid probes. Cancer Genet Cytogenet 1994;74:13–18. [DOI] [PubMed] [Google Scholar]
- 9.McManus AP, Gusterson BA, Pinkerton CR, et al. Diagnosis of Ewing’s sarcoma and related tumours by detection of chromosome 22q12 translocations using fluorescence in situ hybridization on tumour touch imprints. J Pathol 1995;176:137–42. [DOI] [PubMed] [Google Scholar]
- 10.Nagao K, Ito H, Yoshida H, et al. Chromosomal rearrangement t(11;22) in extraskeletal Ewing’s sarcoma and peripheral neuroectodermal tumour analysed by fluorescence in situ hybridization using paraffin-embedded tissue. J Pathol 1997;181:62–6. [DOI] [PubMed] [Google Scholar]
- 11.Kumar S, Pack S, Kumar D, et al. Detection of EWS–Fli-1 fusion in Ewing’s sarcoma/peripheral neuroectodermal tumor by fluorescence in situ hybridization using formalin-fixed paraffin-embedded tissue. Hum Pathol 1999;30:324–30. [DOI] [PubMed] [Google Scholar]
- 12.Gardner LJ, Ayala AG, Monforte HL, et al. Ewing sarcoma/peripheral primitive neuroectodermal tumor: adult abdominal tumors with a Ewing sarcoma gene rearrangement demonstrated by fluorescence in situ hybridization in paraffin sections. Appl Immunohistochem Mol Morphol 2004;12:160–5. [DOI] [PubMed] [Google Scholar]
- 13.Deyrup AT, Althof P, Zhou M, et al. Paraganglioma-like dermal melanocytic tumor: a unique entity distinct from cellular blue nevus, clear cell sarcoma, and cutaneous melanoma. Am J Surg Pathol 2004;28:1579–86. [DOI] [PubMed] [Google Scholar]
- 14.D’Antonio A, Caleo A, Garcia JF, et al. Primary peripheral PNET/Ewing’s sarcoma of the dura with FISH analysis. Histopathology 2004;45:651–4. [DOI] [PubMed] [Google Scholar]
- 15.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987;162:156–9. [DOI] [PubMed] [Google Scholar]
- 16.Giovannini M, Biegel JA, Serra M, et al. EWS–erg and EWS–Fli1 fusion transcripts in Ewing’s sarcoma and primitive neuroectodermal tumors with variant translocations. J Clin Invest 1994;94:489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerald WL, Rosai J, Ladanyi M. Characterization of the genomic breakpoint and chimeric transcripts in the EWS–WT1 gene fusion of desmoplastic small round cell tumor. Proc Natl Acad Sci U S A 1995;92:1028–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonescu CR, Tschernyavsky SJ, Woodruff JM, et al. Molecular diagnosis of clear cell sarcoma: detection of EWS–ATF1 and MITF–M transcripts and histopathological and ultrastructural analysis of 12 cases. J Mol Diagn 2002;4:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis RJ, D’Cruz CM, Lovell MA, et al. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res 1994;54:2869–72. [PubMed] [Google Scholar]
- 20.Clark J, Rocques PJ, Crew AJ, et al. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 1994;7:502–8. [DOI] [PubMed] [Google Scholar]
- 21.Marley EF, Liapis H, Humphrey PA, et al. Primitive neuroectodermal tumor of the kidney—another enigma: a pathologic, immunohistochemical, and molecular diagnostic study. Am J Surg Pathol 1997;21:354–9. [DOI] [PubMed] [Google Scholar]
- 22.Sheaff M, McManus A, Scheimberg I, et al. Primitive neuroectodermal tumor of the kidney confirmed by fluorescence in situ hybridization. Am J Surg Pathol 1997;21:461–8. [DOI] [PubMed] [Google Scholar]
- 23.Ambros IM, Ambros PF, Strehl S, et al. MIC2 is a specific marker for Ewing’s sarcoma and peripheral primitive neuroectodermal tumors. Evidence for a common histogenesis of Ewing’s sarcoma and peripheral primitive neuroectodermal tumors from MIC2 expression and specific chromosome aberration. Cancer 1991;67:1886–93. [DOI] [PubMed] [Google Scholar]
- 24.Arvand A, Denny CT. Biology of EWS/ETS fusions in Ewing’s family tumors. Oncogene 2001;20:5747–54. [DOI] [PubMed] [Google Scholar]
- 25.Gerald WL, Miller HK, Battifora H, et al. Intra-abdominal desmoplastic small round cell tumor. Report of 19 cases of a distinctive type of high-grade polyphenotypic malignancy affecting young individuals. Am J Surg Pathol 1991;15:499–513. [PubMed] [Google Scholar]
- 26.Gerald WL, Rosai J. Desmoplastic small round cell tumor with multiphenotypic differentiation. Zentralbl Pathol 1993;139:141–51. [PubMed] [Google Scholar]
- 27.Ordonez NG, El-Naggar AK, Ro JY, et al. Intra-abdominal desmoplastic small round cell tumor: a light microscopic, immunocytochemical, ultrastructural and flow cytometric study. Hum Pathol 1993;24:850–65. [DOI] [PubMed] [Google Scholar]
- 28.Gerald WL, Ladanyi M, De Alava E, et al. Clinical, pathologic, and molecular spectrum of tumors associated with t(11;22)(p13;q12): desmoplastic small round-cell tumor and its variants. J Clin Oncol 1998;16:3028–36. [DOI] [PubMed] [Google Scholar]
- 29.Ladanyi M, Gerald WL. Fusion of EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res 1994;54:2837–40. [PubMed] [Google Scholar]
- 30.De Alava E, Ladanyi M, Rosai J, et al. Detection of chimeric transcripts in desmoplastic small round cell tumors by RT-PCR. Am J Pathol 1995;147:1584–91. [PMC free article] [PubMed] [Google Scholar]
- 31.Argatoff LH, O’Connell JX, Mathers JA, et al. Detection of the EWS/WT1 gene fusion by reverse transcriptase-polymerase chain reaction in the diagnosis of intra-abdominal desmoplastic small round cell tumor. Am J Surg Pathol 1996;20:406–12. [DOI] [PubMed] [Google Scholar]
- 32.Shen WP, Towne B, Zadeh TM. Cytogenetic abnormalities in an intra-abdominal desmoplastic small round cell tumour. Cancer Genet Cytogenet 1992;64:189–91. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez E, Sreekantaiah C, Gerald W, et al. A recurring translocation, t(11;22)(p13;q12), characterizes intra-abdominal desmoplastic small round cell tumours. Cancer Genet Cytogenet 1993;69:17–21. [DOI] [PubMed] [Google Scholar]
- 34.Roberts P, Burchill SA, Beddow RA, et al. A combined cytogenetic and molecular approach to diagnosis in a case of desmoplastic small round cell tumor with a complex translocation (11;22;21). Cancer Genet Cytogenet 1999;108:19–25. [DOI] [PubMed] [Google Scholar]
- 35.Bridge JA, Borek DA, Neff JR, et al. Chromosomal abnormalities in clear cell sarcoma: implication for histogenesis. Am J Clin Pathol 1990;93:26–31. [DOI] [PubMed] [Google Scholar]
- 36.Peulve P, Michot C, Vannier JP, et al. Clear cell sarcoma with t(12;22)(q13–14;q12). Genes Chromosomes Cancer 1991;3:400–2. [DOI] [PubMed] [Google Scholar]
- 37.Fletcher JA. Translocation (12;22)(q13–14;q12) is a nonrandom aberration in soft-tissue clear-cell sarcoma. Genes Chromosomes Cancer 1992;5:184. [DOI] [PubMed] [Google Scholar]