Abstract
Aims: To investigate the expression of the imprinted oncofetal H19 gene in hepatic metastases derived from a range of human carcinomas and assess its prognostic value with the view of developing a DNA based treatment for such metastases.
Methods: Non-radioactive in situ hybridisation for H19 RNA was performed on paraffin wax embedded sections of liver biopsies or partial hepatectomy specimens, taken from 80 patients with hepatic metastases derived from carcinomas from several medical centres in Israel. The degree of expression was graded qualitatively according to the number of cells expressing H19 and the intensity of staining. The medical files were searched for demographic data and survival times before and after diagnosis of hepatic metastases.
Results: H19 expression was found in the hepatic metastases of 64 of 80 patients. High expression (higher staining grades) of H19 in the metastases was found in 43 of 80 patients. However, H19 expression status in the hepatic metastases did not correlate with either the length of time to development of metastasis or overall survival.
Conclusions: H19 is highly expressed in more than half of hepatic metastases derived from a range of carcinomas. Thus, these metastases may be suitable candidates for H19 DNA based treatment. Further studies are needed to determine whether H19 expression has prognostic value in metastatic liver disease using larger numbers of specific subtypes of primary carcinomas.
Keywords: H19 gene, hepatic metastases (from carcinomas), in situ hybridisation, DNA based therapy, prognostic value
The liver is one of the major sites of cancer metastasis.1 The most common primary malignancies producing hepatic metastases are carcinomas originating in the gastrointestinal tract, breast, and lung, in addition to melanoma, but cancer from any site may spread to the liver.1 This is often an ominous sign, indicating poor prognosis.1 However, it has been shown that in cases of colon and perhaps even breast carcinoma with a limited number of metastases, partial hepatectomy can provide hope for longterm survival.1,2 Other treatment methods for hepatic metastases include hepatic arterial chemotherapy, systemic chemotherapy, chemoembolisation, and several ablative techniques such as ethanol injection, cryoablation, radiofrequency ablation, microwave ablation, interstitial laser photocoagulation, and high intensity focused ultrasound.3–5 All these methods have some, albeit limited, success in extending patient survival or in palliative treatment.3–7 There are also some promising novel therapeutic modalities based on the molecular biology of metastasis in various phases of development.8 These include: antiangiogenesis agents, metalloprotease inhibitors, immunotherapy, and gene therapy based treatment strategies.8
“The aim of our present study was to determine whether H19 is expressed in hepatic metastases from a range of human carcinomas, with a view to establishing the mechanistic basis for developing a DNA based treatment for such metastases”
H19 is an imprinted, maternally expressed oncofetal gene that functions as an RNA molecule.9 H19 resides on chromosome 11p15.5, close to the IGF II gene (encoding insulin-like growth factor II) locus.9
These genes are reciprocally imprinted and their expression is linked in fetal and adult tissues.9 H19 is expressed abundantly in many cancers and only marginally in nearly all normal adult tissues.10 The exact biological function of the H19 RNA product is still unknown. Previous works have shown a tumour suppressor effect, both in vitro11 and in vivo, mainly in Wilms’ tumour.12,13 However, other studies, including our own, have shown H19 overexpression in many cancers (hepatocellular carcinoma,9 testicular cancers,14,15 choriocarcinoma,16,17 oesophageal cancer,18 colorectal cancer,18 ovarian cancer,19 and breast cancer20–22), with or without loss of imprinting. Adriaenssens and colleagues have suggested that H19 plays a part in epithelial–mesenchymal interactions and thus in morphogenesis and/or migration of epithelial cells.23 We have demonstrated H19 upregulation in response to stress, and we have postulated that it has a possible role in promoting cancer progression, angiogenesis, and metastasis.10 Recent studies have shown that changes in methylation of promoter/enhancer sites are the trigger for H19 expression in some cancers.24–28
We have shown that H19 is strongly expressed in over 80% of human bladder carcinomas and that its expression decreases with loss of tumour differentiation. This decrease has prognostic value in predicting early tumour recurrence.29 We also found H19 expression in 60% of endometrial carcinomas, and demonstrated a correlation between expression level and histological grade.30
Although a connection between IGF II expression and the development of liver metastases from colorectal cancers has been shown,31 H19 expression in liver metastases in humans has not been studied.
The aim of our present study was to determine whether H19 is expressed in hepatic metastases from a range of human carcinomas, with a view to establishing the mechanistic basis for developing a DNA based treatment for such metastases. This treatment was shown recently to reduce subcapsular induced liver tumours in a metastatic model of rat CC531 colon carcinoma.32
A secondary aim was to examine the prognostic value of H19 expression in hepatic metastases, in view of previous reports, which showed that H19 expression had prognostic relevance in specific tumours (see above).
We found H19 expression in a large proportion of hepatic metastases, and a high level of expression in over half. However, H19 expression in these metastases did not appear to have prognostic relevance.
MATERIALS AND METHODS
Representative paraffin wax blocks of metastatic carcinoma in the liver were selected from the archives of several pathology departments in Israel including Soroka Medical Center (Be’er-Sheva), Wolfson Medical Center (Holon), and Hadassah Medical Organization (Ein-kerem and Mount Scopus branches, Jerusalem). These blocks were gathered from the pool of routinely prepared blocks from core and wedge liver biopsies and partial hepatectomy specimens taken from 80 patients with hepatic metastases derived from a variety of carcinomas. An exemption from the Helsinki committee at Hadassah Medical Organization was secured.
Non-radioactive in situ hybridisation studies were performed on sections from these blocks using the digoxigenin labelled H19 probe according to previously described methods.9 Placental tissue was used as a positive control and controls for the specificity of in situ hybridisation were used as described previously.9 The stained sections were evaluated with a light microscope (Axioplan2; Carl Zeiss Jena GmbH, Göttingen, Germany) by three experienced pathologists and photographed with a CCD camera (Axiocam, Carl Zeiss Jena GmbH).
The degree of staining in the metastatic carcinoma, the adjacent liver parenchyma, and in the primary carcinoma, if available, was graded as follows: no staining (grade 0), a few cells (< 5%) or weak staining (grade 1), up to half of the cells and moderate staining (grade 2), more than half of the cells and strong staining (grade 3). Grade 1 was considered to be low level H19 expression and grades 2 and 3 were considered to be high level H19 expression.
The demographic and clinical data obtained from the patients’ files included sex, primary tumour type, and, when available, exact dates of clinical events, including diagnosis of metastasis and of primary tumour and of death in the event that death had occurred. Patients’ ages at all available clinical event dates were recorded.
Kaplan–Meier plots and log rank and Breslow tests were used for analysis of metastasis free survival and overall survival in association with H19 expression in the metastases. Statistical analysis was performed using SPSS for Windows (version 12; SPSS Inc, Chicago, Illinois, USA).
RESULTS
We studied 80 patients, 45 of whom were female and 35 were male. Table 1 shows the characteristics of the patients and the distribution of H19 expression in their hepatic metastases.
Table 1.
Summary of characteristics of patients with hepatic metastases derived from a variety of carcinomas and H19 level of expression in paraffin section samples of these metastases
| Characteristic | All patients | No H19 expression | Low level H19 expression | High level H19 expression |
| Mean (SD; N) age at metastasis diagnosis (years) | 60.8 (13.3; 80) | 61.1 (9.33; 16) | 61.4 (17.07; 21) | 60.5 (12.79; 43) |
| Mean (SD; N) age at primary tumour diagnosis (years) | 59.75 (14.9; 54) | 64 (7.12; 9) | 58.6 (18.4; 16) | 59 (14.75; 29) |
| No. patients who died (%) | 57 (71) | 13 (23) | 11 (19) | 33 (58) |
| No. patients who remained alive (%) | 12 (15) | 1 (8) | 5 (42) | 6 (50) |
| Mean (SD; N) age at death (years) | 62.7 (12.3; 49) | 62.7 (9.87; 11) | 68.8 (10.93; 9) | 60.8 (13.2; 29) |
| Sex (%) | ||||
| Female | 45 (56) | 7 (16) | 11 (24) | 27 (60) |
| Male | 35 (44) | 9 (26) | 10 (28) | 16 (46) |
| Type of primary carcinoma (%) | ||||
| Colorectal adenocarcinoma | 37 (46) | 6 (16) | 13 (35) | 18 (49) |
| Breast carcinoma | 5 (6) | 1 | 4 | |
| Pancreatic exocrine adenocarcinoma | 3 (4) | 2 | 1 | |
| Pancreatic endocrine tumour | 2 (2.5) | 2 | ||
| Gastric adenocarcinoma | 3 (4) | 3 | ||
| Parotid gland adenoid cystic carcinoma | 1 (1.25) | 1 | ||
| Duodenal adenocarcinoma | 1 (1.25) | 1 | ||
| Urinary bladder transitional cell carcinoma | 1 (1.25) | 1 | ||
| Oesophageal adenocarcinoma | 1 (1.25) | 1 | ||
| Uncertain or unknown primary | 26 (32.5) | 7 (27) | 6 (23) | 13 (50) |
The average age at primary tumour diagnosis (known for 54 patients) was 59.75 years (SD, 14.9). The average age at metastasis diagnosis was 60.8 years (SD, 13.3). Twelve of the 80 patients were still alive and 57 had died. Survival follow up data were unavailable for 11 patients. The exact age of death was known for 49 of the 57 dead patients. The average age of death in this group was 62.7 years (SD, 12.3).
H19 expression was seen in 64 of the 80 metastases (fig 1). High expression (grades 2 and 3) was seen in 43 of the 80 metastases (fig 1). These proportions are similar when considering hepatic metastases derived from primary colorectal adenocarcinoma alone (H19 expression, 31 of 37; high H19 expression, 18 of 37).
Figure 1.
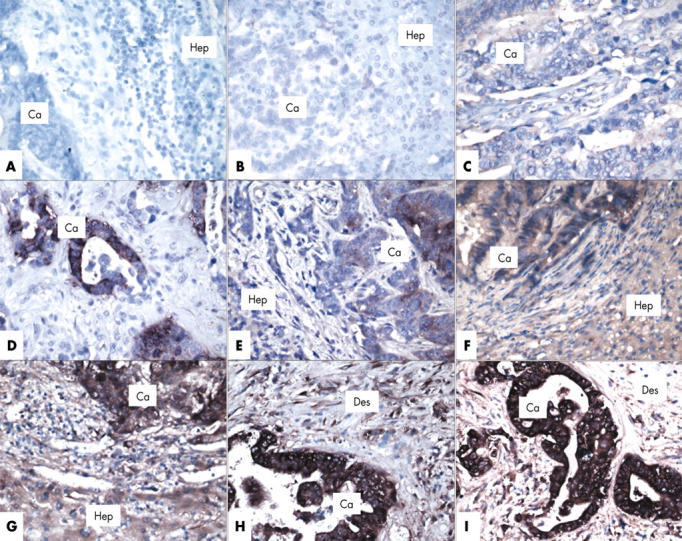
H19 RNA expression in hepatic metastases from carcinomas using a non-radioactive in situ hybridisation method performed on paraffin wax embedded sections. Positive staining is seen as cytoplasmic brown staining. All sections are counterstained with Giemsa. Ca, metastatic carcinoma; Hep, hepatocytes in adjacent liver; Des, desmoplastic reaction. The original primary magnification of all sections is ×400. (A) No staining (grade 0) of the metastatic carcinoma cells or hepatocytes. (B, C) Focal or very weak staining (grade 1) only of the metastatic carcinoma cells (C, section of a primary colonic carcinoma). (D–F) Moderate staining of less than half of the metastatic carcinoma cells (grade 2), and some staining of adjacent hepatocytes (D, section of a primary colonic carcinoma). (G–I) Strong, diffuse staining of the metastatic carcinoma cells (grade 3) and the desmoplastic reaction. Moderate to strong staining of adjacent hepatocytes.
There was strong staining (grades 2 or 3) in the adjacent liver parenchyma in 26 of the 80 metastases (fig 1).
H19 staining of the primary tumour was also available for 13 patients (fig 1). In 11 of these cases, the staining grades were similar to those in the metastases.
The exact time course from primary tumour diagnosis to liver metastasis (metastasis free survival) was known for 54 of the 80 patients. The exact survival period from primary tumour diagnosis to death or to most recent follow up (overall survival) was known for 38 of the 80 patients.
Of the 54 patients in the first group, 29 had high level (grades 2 or 3) H19 expression in their metastases, and a mean metastasis free survival of 19.18 months (SE, 8.57; 95% confidence interval (CI), 2.39 to 35.98; median, 0.00). Sixteen patients had low level H19 expression (grade 1) in their metastases, and a mean metastasis free survival of 6.71 months (SE, 4.44; 95% CI, 0.00 to 15.4; median, 0.00). Nine patients had H19 negative metastases, and a mean metastasis free survival of 0.25 months (SE, 0.18; 95% CI, 0.00 to 0.59; median, 0.00). Although Kaplan–Meier plots (fig 2A) show differences in cumulative survival between the three groups, the correlation between the level of H19 expression in the metastases and metastasis free survival is not significant either by the log rank test (p = 0.12) or by the Breslow test (p = 0.42).
Figure 2.
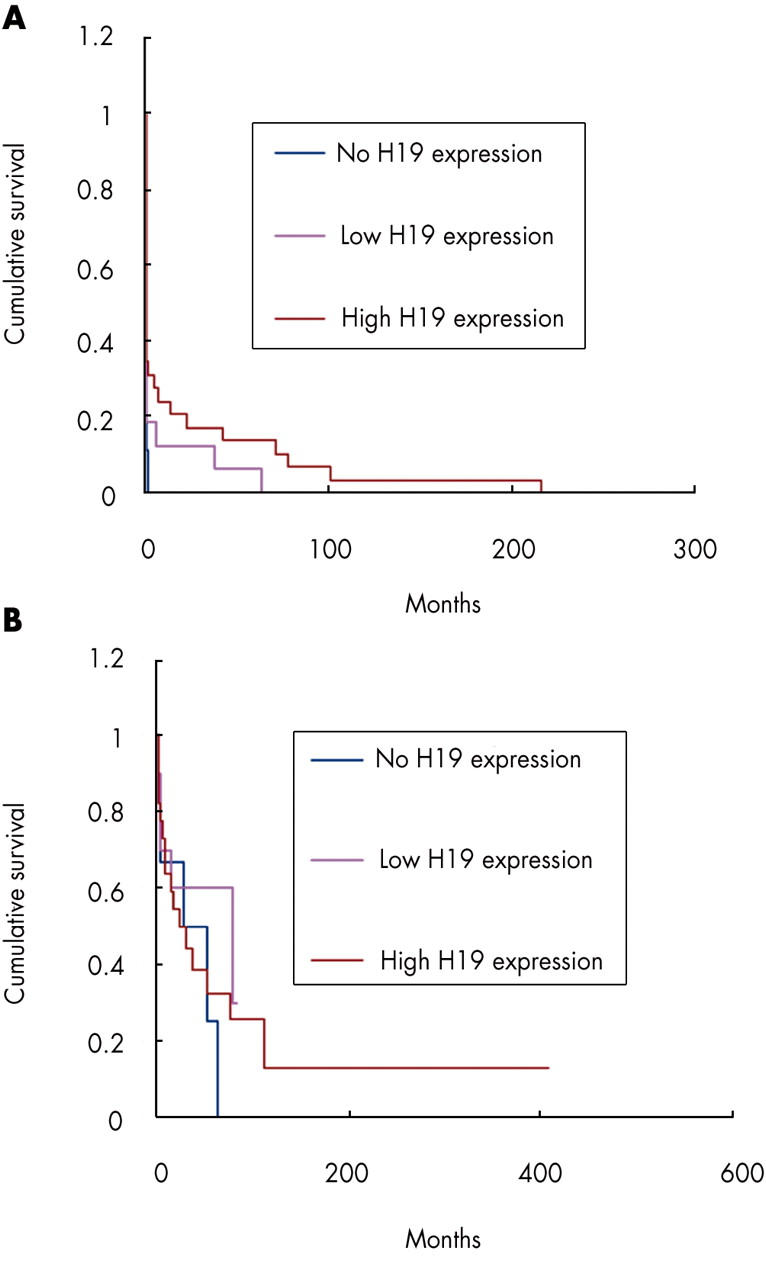
The correlation between survival and H19 expression in hepatic metastases derived from carcinomas represented by Kaplan–Meier plots. (A) Survival from primary tumour diagnosis to metastasis diagnosis (metastasis free survival). p = 0.12 by log rank test and 0.42 by Breslow test. (B) Survival from primary tumour diagnosis to death or to last follow up (overall survival). p = 0.61 by log rank test and 0.82 by Breslow test.
Of the 38 patients in the second group, 22 had high level H19 expression in their metastases, a mean overall survival of 83.61 months (SE, 36.40; 95% CI, 12.27 to 154.95 (limited to 409.11)), and a median overall survival of 24.87 months (SE, 11.75; 95% CI, 1.84 to 47.90). Ten patients had low level H19 expression in their metastases, a mean overall survival of 51.97 months (SE, 11.73; 95% CI, 28.97 to 74.97 (limited to 83.85)), and a median overall survival of 80.26 months (SE, 48.04; 95% CI, 0.00 to 174.42). Six patients had H19 negative metastases, a mean overall survival of 34.52 months (SE, 11.65; 95% CI, 11.69 to 57.36), and a median overall survival of 28.06 months (SE, 24.12; 95% CI, 0.00 to 75.34). Although Kaplan–Meier plots (fig 2B) show differences in cumulative survival between the three groups, the correlation between the level of H19 expression in the metastases and overall survival is again not significant either by the log rank test (p = 0.61) or by the Breslow test (p = 0.82).
DISCUSSION
H19, an imprinted, maternally expressed oncofetal gene that functions as an RNA molecule,9 is expressed abundantly in many cancers.10 Its exact biological role is largely unknown, but it is probably involved in promoting cancer progression, angiogenesis, and metastasis.10
We were interested in H19 expression in hepatic metastases derived from carcinomas, because the liver is one of the major sites of cancer metastasis.1 As described, we evaluated H19 expression in paraffin wax embedded sections of 80 metastatic carcinomas in the liver, using non-radioactive in situ hybridisation. We considered hepatic metastasis as a liver disease,1 and thus we did not try to select the samples deliberately according to the exact statistical distribution of primary carcinomas. Although colorectal carcinoma is the main primary carcinoma to metastasise to the liver,1 and is the main primary tumour in our group (37 of 80; table 1), the distribution of primary tumours is probably biased because most partial hepatectomies are performed for metastatic colorectal carcinomas. In addition, most of the primary tumours in the second largest group (uncertain or unknown primary; table 1) are probably colorectal carcinomas.
“We conclude that the diphtheria toxin A chain–H19 vector appears to be a very good candidate for a patient oriented DNA based treatment for metastatic carcinoma in the liver”
H19 expression was seen in 64 of the 80 metastases, with high level expression (grade 2–3) in 43. This is similar to its expression in some primary carcinomas.9,14,18,19,21,29,30 Indeed, H19 expression was seen in most subgroups of primary tumours (table 1). These results imply that metastatic carcinomas in the liver originating from various primary tumours express H19, thus establishing the mechanistic basis for developing a DNA based treatment targeted to such metastases, without considering the primary carcinoma.
H19 expression did not appear to have prognostic value in terms of metastasis free survival (fig 2A) or overall survival (fig 2B). However, the results might have been influenced by lack of accurate data for many patients. Ideally, tests of prognostic relevance should be conducted on a larger population, with stratification according to sex, age, and primary tumour subtype.
With regard to harnessing H19 expression in hepatic metastases for specific DNA based antimetastatic treatment, we have developed vectors carrying the diphtheria toxin A chain gene under the control of IGF-II promoters 3 and 4, and H19 regulatory sequences, and have shown that these vectors selectively destroy tumour cells and inhibit tumour growth in vitro and in vivo33,34—for example, in a rat model of urinary bladder carcinoma,35 in human transitional cell carcinoma,35 and in reducing subcapsular induced liver tumours in a metastatic model of rat CC531 colon carcinoma.32 Because we have shown here that H19 is abundantly expressed in hepatic metastases derived from a range of carcinomas, taken together with the above findings, we conclude that the diphtheria toxin A chain–H19 vector appears to be a very good candidate for a patient oriented DNA based treatment for metastatic carcinoma in the liver. Our results in treating patients with transitional cell carcinomas35 and one patient with a liver metastasis originating from colon carcinoma (unpublished data; A Hochberg, 2004) suggest that this is a relatively safe treatment.
In many of the metastases, there was a significant desmoplastic reaction, which mostly showed similar staining to the adjacent metastatic carcinoma. This is not surprising because it is known that H19 is expressed within both epithelial and stromal components of invasive adenocarcinomas.20–22 The staining of the desmoplastic reaction might also be related to the alleged contribution of H19 to epithelial–mesenchymal interactions.23
In approximately a third of the cases, there was significant staining (grades 2 or 3) in the adjacent liver. Expression of growth factors (such as IGF-I and IGF-II) and angiogenesis promoting factors (such as angiopoietin 2) by host organs with metastases is a well recognised phenomenon.36 IGF-II overexpression by hepatocytes adjacent to liver metastases derived from colorectal carcinomas suggests its possible contribution to the development of such metastases.31 Moreover, H19 is coupled to IGF-II and probably plays a role in angiogenesis.10,23 This may explain its expression in hepatocytes adjacent to metastases. The expression of H19 in the adjacent liver tissue is unlikely to pose a problem for the proposed DNA based treatment (see above) because to destroy a tumour efficiently, its microenvironment should also be destroyed,36 and the treatment is highly specific and should be injected directly into the metastasis (unpublished data; A Hochberg, 2004).
Take home messages.
H19 is an imprinted, maternally expressed oncofetal gene expressed abundantly in many cancers and only marginally in normal adult tissues
We found that H19 is highly expressed in more than half of hepatic metastases derived from a range of carcinomas
Thus, these metastases may be suitable candidates for H19 DNA based treatment, such as vectors carrying the diphtheria toxin A chain gene under the control of the H19 promoter
Further studies are needed to determine whether H19 expression has prognostic value in metastatic liver disease using larger numbers of specific subtypes of primary carcinomas
In a small number of the patients, H19 staining of the primary tumour was also available, and in most of these cases, the staining intensity was similar to that in the metastasis. Although our results require verification in larger scale studies, it is possible that knowledge of the H19 expression profile in the primary tumour may eventually suffice in the clinical decision of whether to use an H19 based treatment against its hepatic metastases.
Acknowledgments
To Dr K Meir and Dr E Pikarsky from the Pathology Department in Hadassah Medical Organization, Jerusalem, Israel for their essential advice and contribution.
Abbreviations
CI, confidence interval
IGF, insulin-like growth factor
REFERENCES
- 1.Ishac KG, Goodman ZD, Stocker JT, eds. Metastatic tumors. In: Tumors of the liver and intrahepatic bile ducts. Atlas of tumor pathology, 3rd series, fascicle 31. Washington DC: Armed Forces Institute of Pathology, 2001:343–9.
- 2.Elias D, Maisonnette F, Druet-Cabanac M, et al. An attempt to clarify indications for hepatectomy for hepatic metastases from breast cancer. Am J Surg 2003;185:158–64. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya R, Rao S, Kowdley KV. Liver involvement in patients with solid tumors of non-hepatic origin. Clin Liver Dis 2002;6:1033–43. [DOI] [PubMed] [Google Scholar]
- 4.Erce C, Parks RW. Interstitial ablative techniques for hepatic tumors. Br J Surg 2003;90:272–89. [DOI] [PubMed] [Google Scholar]
- 5.Bonetti A. Hepatic artery infusion for hepatic metastases from colorectal cancer [commentary]. Lancet 2003;361:358–9. [DOI] [PubMed] [Google Scholar]
- 6.Adam R, Lucidi V, Bismuth H. Hepatic colorectal metastases: methods of improving resectability. Surg Clin North Am 2004;84:659–71. [DOI] [PubMed] [Google Scholar]
- 7.Khatri VP, McGahan J. Non-resection approaches for colorectal hepatic metastases. Surg Clin North Am 2004;84:587–606. [DOI] [PubMed] [Google Scholar]
- 8.Allendorf J, Ippagunta N, Emond J. Management of hepatic metastases: new horizons for biologically based therapy. J Surg Res 2004;117:144–53. [DOI] [PubMed] [Google Scholar]
- 9.Ariel I, Miao HQ, Ji XR, et al. Imprinted H19 oncofetal RNA is a candidate tumor marker for hepatocellular carcinoma. Mol Pathol 1998;51:21–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayesh S, Matouk I, Schneider T, et al. Possible physiological role of H19 RNA. Mol Carcinog 2002;35:63–74. [DOI] [PubMed] [Google Scholar]
- 11.Hao Y, Crenshaw T, Moulton T, et al. Tumor-suppressor activity of H19 RNA. Nature 1993;365:764–7. [DOI] [PubMed] [Google Scholar]
- 12.Moulton T, Crenshaw T, Hao Y, et al. Epigenetic lesions at the H19 locus in Wilms’ tumor patients. Nat Genet 1994;7:440–7. [DOI] [PubMed] [Google Scholar]
- 13.Taniguchi T, Sullivan MJ, Ogawa O, et al. Epigenetic changes encompassing the IGF2/H19 locus associated with relaxation of IGF2 imprinting and silencing of H19 in Wilms tumor. Proc Natl Acad Sci U S A 1995;92:2159–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Gurp RJ, Oosterhuis JW, Kalscheuer V, et al. Biallelic expression of the H19 and IGF2 genes in human testicular germ cell tumors. J Natl Cancer Inst 1994;86:1070–5. [DOI] [PubMed] [Google Scholar]
- 15.Verterk AJMH, Ariel I, Dekker MC, et al. Unique expression patterns of H19 in human testicular cancers of different etiology. Oncogene 1997;14:95–107. [DOI] [PubMed] [Google Scholar]
- 16.Ariel I, Lustig O, Oyer CE, et al. Relaxation of imprinting in trophoblastic disease. Gynecol Oncol 1994;53:212–19. [DOI] [PubMed] [Google Scholar]
- 17.Lustig-Yariv O, Schulze E, Komitowski D, et al. The expression of the imprinted genes H19 and IGF-2 in choriocarcinoma cell lines. Is H19 a tumor suppressor gene? Oncogene 1997;15:169–77. [DOI] [PubMed] [Google Scholar]
- 18.Hibi K, Nakamura H, Hirai A, et al. Loss of H19 imprinting in esophageal cancer. Cancer Res 1996;56:480–2. [PubMed] [Google Scholar]
- 19.Tanos V, Prus D, Ayesh S, et al. Expression of the imprinted H19 oncofetal RNA in epithelial ovarian cancer. Eur J Obstet Gynecol Reprod Biol 1999;85:7–11. [DOI] [PubMed] [Google Scholar]
- 20.Dugimont T, Curgy JJ, Wernert N, et al. The H19 gene is expressed within both epithelial and stromal components of human invasive adenocarcinomas. Biol Cell 1995;85:117–24. [DOI] [PubMed] [Google Scholar]
- 21.Adriaenssens E, Dumont L, Lottin S, et al. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of P53 and Ki-67 expression. Am J Pathol 1998;153:1597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lottin S, Adriaenssens E, Dupressoir T, et al. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogen 2002;23:1885–95. [DOI] [PubMed] [Google Scholar]
- 23.Adriaenssens E, Lottin S, Berteaux N, et al. Cross talk between mesenchyme and epithelium increases H19 gene expression during scattering and morphogenesis of epithelial cells. Exp Cell Res 2002;275:215–29. [DOI] [PubMed] [Google Scholar]
- 24.Lynch CA, Tycko B, Bestor TH, et al. Reactivation of a silenced H19 gene in human rhabdomyosarcoma by demethylation of DNA but not by histone hyperacetylation. Mol Cancer 2002;1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanduri C, Kanduri M, Liu L, et al. The kinetics of deregulation of expression by de novo methylation of the H19 imprinting control region in cancer cells. Cancer Res 2002;62:4545–8. [PubMed] [Google Scholar]
- 26.Ulaner GA, Vu TH, Li T, et al. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum Mol Genet 2003;12:535–49. [DOI] [PubMed] [Google Scholar]
- 27.Ng A, Tang JP, Goh CHK, et al. Regulation of the H19 imprinting gene expression in human nasopharyngeal carcinoma by methylation. Int J Cancer 2003;104:179–87. [DOI] [PubMed] [Google Scholar]
- 28.Vernucci M, Cerrato F, Pedone PV, et al. Developmentally regulated functions of the H19 differentially methylated domain. Hum Mol Genet 2004;13:353–61. [DOI] [PubMed] [Google Scholar]
- 29.Ariel I, Sughayer M, Fellig Y, et al. The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol Pathol 2000;53:320–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanos V, Ariel I, Prus D, et al. H19 and IGF2 gene expression in human normal, hyperplastic and malignant endometrium. Int J Gynecol Cancer 2004;14:521–5. [DOI] [PubMed] [Google Scholar]
- 31.Kawamoto K, Onodera H, Kan S, et al. Possible paracrine mechanism of insulin-like-growth-factor-2 in the development of liver metastases from colorectal carcinoma. Cancer 1999;85:18–25. [DOI] [PubMed] [Google Scholar]
- 32.Ohana P, Schachter P, Ayesh B, et al. Regulatory sequences of H19 and IGF2 genes in DNA based therapy of colorectal rat hepatic metastases. J Gene Med 2005;7:366–74. [DOI] [PubMed] [Google Scholar]
- 33.Ohana P, Bibi O, Matouk I, et al. Use of H19 regulatory sequences for targeted gene therapy in cancer. Int J Cancer 2002;98:645–50. [DOI] [PubMed] [Google Scholar]
- 34.Ayesh B, Matouk I, Ohana P, et al. Inhibition of tumor growth by DT-A expressed under the control of IGF2 P3 and P4 promoter sequences. Mol Ther 2003;7:535–41. [DOI] [PubMed] [Google Scholar]
- 35.Ohana P, Gofrit O, Ayesh S, et al. Regulatory sequences of the H19 gene in DNA based therapy of bladder cancer. Gene Ther Mol Biol 2004;8:181–92. [Google Scholar]
- 36.Bogenrieder T, Herlyn M. Axis of evil: molecular mechanisms of cancer metastasis. Oncogene 2003;22:6524–36. [DOI] [PubMed] [Google Scholar]


