Abstract
Allelic losses on chromosome 1p are frequent in hepatocellular carcinoma (HCC), suggesting the presence of a tumour suppressor gene in this region. The gene for peroxiredoxin 1 (PRDX1), an antioxidant enzyme, has been mapped to 1p34.1. Mice lacking PRDX1 develop HCC with high frequency. Because oxidative stress has been implicated in the pathogenesis of HCC, this study was designed to determine whether the PRDX1 gene is mutated in human HCC using loss of heterozygosity (LOH) analysis, polymerase chain reaction/denaturing gradient gel electrophoresis, and DNA sequencing. LOH of at least one of four microsatellite markers within 0.8 Mb of the PRDX1 gene was seen in three of 34 informative HCCs, but no mutations or polymorphisms in the translated exons 2–6 of the PRDX1 gene were found. These results suggest that genetic alterations of the PRDX1 locus are rare events in human HCC, indicating that other genes on chromosome 1p contribute to liver carcinogenesis.
Keywords: hepatocellular carcinoma, PRDX1, peroxiredoxin, chromosome 1p34, oxidative damage
Oxidative stress caused by free radicals has been implicated in the pathogenesis of many cancers.1 Free radicals, such as reactive oxygen species (ROS), may cause mutations in cancer related genes or directly alter the functions of proteins regulating DNA repair, cell cycle progession, and apoptosis. Oxidative stress has been linked to conditions that are associated with an increased risk of hepatocarcinogenesis, such as viral hepatitis, alcoholic liver disease, toxic liver injury, and fibrosis.2,3 Furthermore, hepatocellular carcinoma (HCC) shows evidence of oxidative DNA damage, as demonstrated by the formation of 8-hydroxy-2′-deoxyguanosine.4,5 These observations suggest that ROS induced oxidative damage contributes to the pathogenesis of HCC.
“Mice lacking Prdx1 develop severe haemolytic anaemia and various types of malignancies, including hepatocellular carcinoma, which suggests that this protein functions as a tumour suppressor”
Several studies using karyotyping, comparative genomic hybridisation, and loss of heterozygosity (LOH) analysis have shown that loss of chromosome 1p is a recurrent genetic aberration in human HCC.6 Chromosomal losses most often involve the distal regions of chromosome 1p, with the 1p34–36 region involved in up to 75% of HCCs. The shortest regions of overlap on chromosome 1p have recently been mapped to 1p36.22–p36.13 and 1p36.32–p36.22.7,8 So far, three candidate tumour suppressor genes on the 1p36 region, the PRDM2 (RIZ), RUNX3, and TP73 genes, have been studied in HCC, but no mutations have been detected.8,9,10
Peroxiredoxin 1 (PRDX1)—also designated MSP23, OSF-3, or PAG—is an intracellular antioxidant protein with thioredoxin dependent peroxidase activity, which protects cells against oxidative stress by inactivating ROS, and is encoded by a single gene on chromosome 1p34.1.11 PRDX1 is highly expressed in the liver and was localised to hepatocytes and Kupffer cells in the rat.12,13 Mice lacking Prdx1 develop severe haemolytic anaemia and various types of malignancies, including HCC, which suggests that this protein functions as a tumour suppressor.14 However, PRDX1 gene mutations have not been identified in human tumours.
Because distal chromosome 1p deletions are frequent in human HCC and Prdx1 mutant mice develop HCC with high frequency, we tested the hypothesis that mutations or polymorphisms of the PRDX1 gene may also contribute to the carcinogenesis of human HCC.
MATERIALS AND METHODS
Tumours, DNA extraction
Formalin fixed, paraffin wax embedded tumour samples of 36 HCCs and corresponding non-tumorous liver tissue were drawn from the files of the department of pathology, University Hospital Zürich, Switzerland, covering a period of 11 years (1992 to 2003). Tumour and non-tumorous liver tissue was macrodissected from 10 μm sections and DNA was extracted as described previously.15
LOH analysis
LOH was investigated using four polymorphic markers of the 1p34.1 region located within 0.8 Mb of the PRDX1 locus (telomeric: D1S421; centromeric: D1S2677, D1S451, and D1S2802). Microsatellite markers were amplified from DNA extracted from HCC and corresponding non-tumorous liver tissue. Standard polymerase chain reaction (PCR) was performed using conditions and fluorescent labelled primers as obtained from the Ensembl Genome Browser (www.ensembl.org). PCR products were analysed on an automated DNA sequencer (model 373A; Applied Biosystems, Foster City, California, USA) using Gene Scan software (Applied Biosystems). Cases were classified as informative when two distinct alleles of similar intensity were found in the normal DNA. LOH was defined as present when an allele peak signal from tumour DNA was reduced by ⩾ 50% compared with the corresponding non-tumorous liver DNA.
PCR, denaturing gradient gel electrophoresis, and DNA sequencing
Primers amplifying the translated exons 2–6 were designed based on the reported genomic sequence of the human PRDX1 gene (ENSG00000117450; www.ensembl.org): exon 2F, 5′-GTA GGT GAA GGC TGC TGG TT-3′; exon 2R, 5′-CAT CTC AAT GAG CCC AGT GTT-3′; exon 3F, 5′-TGC TAA CCA TGA CTC CGA TTT-3′; exon 3R, 5′-TTC ACA TGC CAA ATA GAC CAA-3′; exon 4F, 5′-GAA AGG GGA AAG AGG AAT GC-3′; exon 4R, 5′-GGC TTT CAG CCA ACT GGA TA-3′; exon 5F, 5′-AAC CTG TAT TGC TTT TCC TTT CA-3′; exon 5R, 5′-CAC CCC TTC ATA CCA CCA CT-3′; exon 6F, 5′-ATG GTG GTG CAT ACC ATT GA-3′; and exon 6R, 5′-GCA GCC TGG CAC TAA AAC AG-3′. PCR amplification, denaturing gradient gel electrophoresis, and cycle sequencing of PCR products were performed as described previously.15 Sequences were compared with the reported genomic sequence of the PRDX1 gene (ENSG00000117450; www.ensembl.org).
RESULTS AND DISCUSSION
Three lines of evidence encouraged us to investigate genetic alterations of the PRDX1 gene in human HCC. First, the PRDX1 gene maps to the distal region of chromosome 1 (1p34.1), a region of frequent LOH in HCC. So far, three candidate tumour suppressor genes at the 1p36 region, the PRDM2 (RIZ), RUNX3, and TP73 genes, have been studied in HCC and no mutations have been detected, leaving the putative tumour suppressor gene(s) in this region unidentified.8,9,10 Second, HCCs show evidence of oxidative damage, suggesting that defects in ROS scavenging systems may contribute to the initiation and/or promotion of HCC.4,5 Because the PRDX1 protein participates in the antioxidant defence of hepatocytes, PRDX1 inactivation may enhance oxidative stress and thereby contribute to hepatocarcinogenesis. Third, HCCs occur with high penetrance in Prdx1 mutant mice, suggesting a tumour suppressor function for PRDX1 in hepatocyte progenitor cells.14
Take home messages.
The peroxiredoxin 1 gene (PRDX1) is a good candidate for the tumour suppressor gene implicated in the pathogenesis of hepatocellular carcinoma (HCC)
However, loss of heterozygosity of four markers within 0.8 Mb of the PRDX1 gene was seen in only three of 34 informative HCCs and no mutations or polymorphisms in the translated exons 2–6 of the PRDX1 gene were found
These results suggest that genetic alterations of the PRDX1 locus are rare events in human HCC, indicating that other genes on chromosome 1p contribute to liver carcinogenesis
“We cannot exclude the possibility that PRDX1 inactivation by epigenetic mechanisms, such as promoter hypermethylation, may contribute to hepatocarcinogenesis”
To determine whether genetic alterations of the PRDX1 gene are involved in human hepatocarcinogenesis, we first analysed this series of HCCs for LOH of four polymorphic microsatellite markers that are located telomeric and centromeric of the PRDX1 locus on chromosome 1p34. Thirty four of 36 HCCs were informative for at least one of the four markers. Allelic loss of at least one microsatellite marker was seen in three of the 34 informative HCCs. Allelic loss of D1S421, D1S2677, D1S451, and D1S2802 was found in one of 14, two of 21, one of 10, and three of 26 informative cases, respectively (fig 1; table 1).
Figure 1.
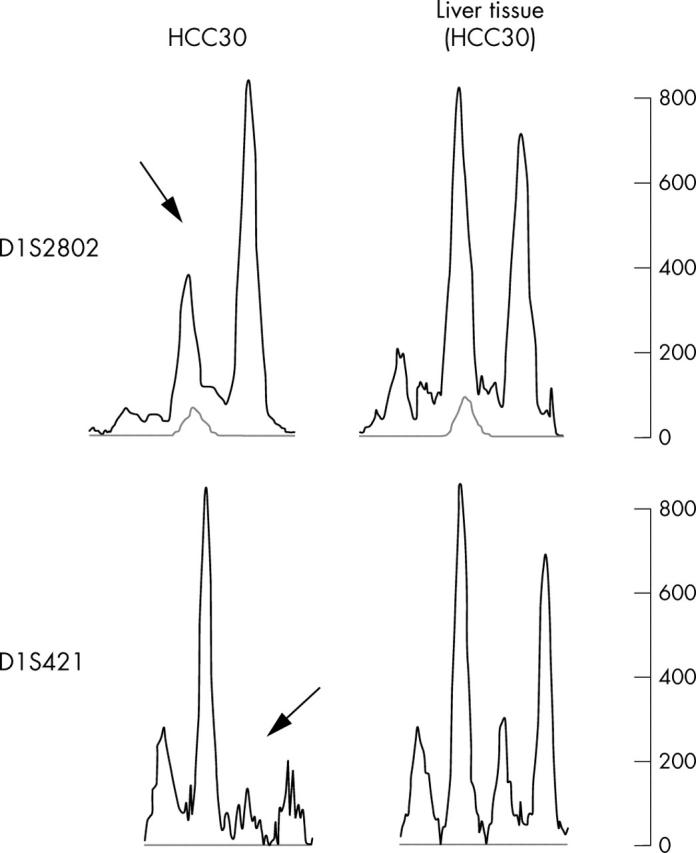
Loss of heterozygosity (LOH) analysis of a moderately differentiated hepatocellular carcinoma (HCC30) with allelic losses (arrows) of the microsatellite markers D1S421 and D1S2802, which are located telomeric and centromeric of the PRDX1 locus, respectively.
Table 1.
Loss of heterozygosity analysis of the 1p34.1 region in primary hepatocellular carcinomas
| Patient | Age/Sex | Aetiology | Tumour size (cm) | Grading | D1S421 | D1S2802 | D1S451 | D1S2677 | Mutation |
| HCC1 | 76/M | HCV, cirrhosis | 4.2 | G1 | NI | ROH | ROH | NI | None* |
| HCC7 | 67/M | No cirrhosis | 17 | G2 | NI | ROH | NI | ROH | None* |
| HCC8 | 55/F | HBV, cirrhosis | 5.5 | G2 | NI | ROH | NI | ROH | None* |
| HCC9 | 51/F | HBV, fibrosis | 2.4 | G3 | NI | ROH | NI | ROH | None* |
| HCC10 | 80/F | HCV, cirrhosis | 4.2 | G2 | ROH | ROH | ROH | ROH | None* |
| HCC11 | 62/M | Haemochromatosis, fibrosis | 10.5 | G2 | ROH | ROH | NI | NI | None |
| HCC12 | 48/M | HCV, cirrhosis | 2.5 | G3 | NI | ROH | NI | ROH | None* |
| HCC13 | 82/M | No cirrhosis | 7.0 | G2 | NI | NI | NI | ROH | None* |
| HCC14 | 69/F | HBV, fibrosis | 8.5 | G2 | NI | ROH | NI | ROH | None |
| HCC15 | 71/M | No cirrhosis | 1.6 | G2 | NI | ROH | NI | NI | None |
| HCC16 | 81/F | No cirrhosis | 18.0 | G1 | ROH | NI | NI | NI | None* |
| HCC17 | 64/M | HBV, cirrhosis | 2.1 | G2 | NI | NI | NI | ROH | None |
| HCC18 | 48/M | HCV, cirrhosis | 13.0 | G2 | ROH | ROH | NI | NI | None |
| HCC19 | 56/M | Alcohol, no cirrhosis | 2.1 | G1 | NI | ROH | ROH | NI | None |
| HCC20 | 49/M | HBV, cirrhosis | 10.0 | G2 | ROH | ROH | ROH | ROH | None |
| HCC21 | 37/F | No cirrhosis | 5.0 | G2 | ROH | ROH | NI | ROH | None |
| HCC22 | 46/M | HCV, cirrhosis | 6.0 | G2 | NI | NI | NI | ROH | None |
| HCC23 | 70/M | HCV, cirrhosis | 2.2 | G1 | NI | ROH | NI | ROH | None* |
| HCC24 | 67/M | cirrhosis | 5.0 | G2 | NI | ROH | NI | ROH | None* |
| HCC25 | 65/F | Alcohol, cirrhosis | 3.8 | G2 | NI | LOH | NI | LOH | None* |
| HCC27 | 36/M | HBV, alcohol, cirrhosis | 8.0 | G3 | ROH | ROH | NI | NI | None |
| HCC28 | 63/M | Alcohol, cirrhosis | 3.5 | G2 | NI | NI | NI | NI | None |
| HCC29 | 56/M | Alcohol, no cirrhosis | 4.0 | G2 | NI | NI | NI | NI | None* |
| HCC30 | 65/F | No cirrhosis | 11.0 | G1 | LOH | LOH | NI | LOH | None* |
| HCC31 | 51/M | HCV, cirrhosis | 2.5 | G2 | NI | ROH | NI | ROH | None* |
| HCC33 | 41/F | Alagille syndrome, cirrhosis | 6.5 | G3 | NI | NI | ROH | ROH | None* |
| HCC34 | 75/M | No cirrhosis | 5.0 | G2 | NI | ROH | ROH | NI | None |
| HCC35 | 23/F | HBV, cirrhosis | 18.0 | G2 | NI | LOH | LOH | NI | None* |
| HCC36 | 57/M | HBV, cirrhosis | 6.5 | G2 | ROH | ROH | NI | ROH | None |
| HCC37 | 61/F | HCV, fibrosis | 4.2 | G2 | NI | ROH | ROH | ROH | None |
| HCC38 | 58/ M | HCV, cirrhosis | 2.9 | G2 | ROH | ROH | NI | NI | None |
| HCC39 | 72/M | Cirrhosis | 6.5 | G2 | ROH | NI | NI | NI | None |
| HCC40 | 56/M | Haemochromatosis, fibrosis | 4.8 | G2 | ROH | NI | NI | NI | None |
| HCC41 | 54/M | HCV, fibrosis | 4.0 | G2 | ROH | ROH | ROH | NI | None |
| HCC42 | 69/M | Cirrhosis | 4.5 | G2 | ROH | ROH | ROH | ROH | None |
| HCC43 | 58/M | Alcohol, cirrhosis | 7.5 | G3 | NI | NI | NI | ROH | None |
*PCR products have also been sequenced.
Four microsatellite markers from the 1p34.1 region (D1S421, D1S2677, D1S451, and D1S2802), which are located centromeric and telomeric of the PRDX1 gene, were used.
HBV, hepatitis B virus; HCV, hepatitis C virus; LOH, loss of heterozygosity; ROH, retention of heterozygosity.
Our results agree with previous studies that also reported LOH at 1p34 in HCC, although with higher frequency (29%,16 48–71%,17 and 15%18). These discrepancies may result from the use of different microsatellite markers and may indicate that LOH on chromosome 1p occurs distally of 1p34. Mutation analysis by PCR/denaturing gradient gel electrophoresis of the translated exons 2–6 of the PRDX1 gene did not reveal band shift variants in the 36 HCCs. Subsequent DNA sequencing of exons 2–6 of 15 HCCs, including those with LOH at chromosome 1p34.1, revealed wild-type sequences.
We conclude that genetic alterations of the PRDX1 gene are rare in HCC and that PRDX1 is unlikely to be the target tumour suppressor gene of LOH on 1p. However, we cannot exclude the possibility that PRDX1 inactivation by epigenetic mechanisms, such as promoter hypermethylation, may contribute to hepatocarcinogenesis.
Acknowledgments
We thank H Moch for critical reading of the manuscript. This work was partly supported by the Gebert Rüf Foundation.
Abbreviations
LOH, loss of heterozygosity
PCR, polymerase chain reaction
PRDX1, peroxiredoxin 1
ROS, reactive oxygen species
REFERENCES
- 1.Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer 2003;3:276–85. [DOI] [PubMed] [Google Scholar]
- 2.Shimoda R, Nagashima M, Sakamoto M, et al. Increased formation of oxidative DNA damage, 8-hydroxydeoxyguanosine, in human livers with chronic hepatitis. Cancer Res 1994;54:3171–72. [PubMed] [Google Scholar]
- 3.Parola M, Robino G. Oxidative stress-related molecules and liver fibrosis. J Hepatol 2001;35:297–306. [DOI] [PubMed] [Google Scholar]
- 4.Seki S, Kitada T, Sakaguchi H, et al. Pathological significance of oxidative cellular damage in human alcoholic liver disease. Histopathology 2003;42:365–71. [DOI] [PubMed] [Google Scholar]
- 5.Ichiba M, Maeta Y, Mukoyama T, et al. Expression of 8-hydroxy-2′-deoxyguanosine in chronic liver disease and hepatocellular carcinoma. Liver Int 2003;23:338–45. [DOI] [PubMed] [Google Scholar]
- 6.Suriawinata A, Xu R. An update on the molecular genetics of hepatocellular carcinoma. Semin Liver Dis 2004;24:77–88. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura T, Nishida N, Itoh T, et al. Discrete breakpoint mapping and shortest region of overlap of chromosome arm 1q gain and 1p loss in human hepatocellular carcinoma detected by semiquantitative microsatellite analysis. Genes Chromosomes Cancer 2005;42:34–43. [DOI] [PubMed] [Google Scholar]
- 8.Fang W, Piao Z, Simon D, et al. Mapping of a minimal deleted region in human hepatocellular carcinoma to 1p36.13–p36.23 and mutational analysis of the RIZ (PRDM2) gene localized to the region. Genes Chromosomes Cancer 2000;28:269–75. [PubMed] [Google Scholar]
- 9.Mihara M, Nimura Y, Ichimiya S, et al. Absence of mutation of the p73 gene localized at chromosome 1p36.3 in hepatocellular carcinoma. Br J Cancer 1999;79:164–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao WH, Liu WW. Hemizygous deletion and hypermethylation of RUNX3 gene in hepatocellular carcinoma. World J Gastroenterol 2004;10:376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood ZA, Schroder E, Robin Harris J, et al. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 2003;28:32–40. [DOI] [PubMed] [Google Scholar]
- 12.Lee TH, Yu SL, Kim SU, et al. Characterization of mouse peroxiredoxin I genomic DNA and its expression. Gene 1999;239:243–50. [DOI] [PubMed] [Google Scholar]
- 13.Immenschuh S, Baumgart-Vogt E, Tan M, et al. Differential cellular and subcellular localization of heme-binding protein 23/peroxiredoxin I and heme oxygenase-1 in rat liver. J Histochem Cytochem 2003;51:1621–31. [DOI] [PubMed] [Google Scholar]
- 14.Neumann CA, Krause DS, Carman CV, et al. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature 2003;424:561–5. [DOI] [PubMed] [Google Scholar]
- 15.Mihic-Probst D, Perren A, Schmid S, et al. Absence of BRAF gene mutations differentiates Spitz nevi from malignant melanoma. Anticancer Res 2004;24:2415–18. [PubMed] [Google Scholar]
- 16.Sun M, Eshleman JR, Ferrell LD, et al. An early lesion in hepatic carcinogenesis: loss of heterozygosity in human cirrhotic livers and dysplastic nodules at the 1p36–p34 region. Hepatology 2001;33:1415–24. [DOI] [PubMed] [Google Scholar]
- 17.Leung TH, Wong N, Lai PB, et al. Identification of four distinct regions of allelic imbalances on chromosome 1 by the combined comparative genomic hybridization and microsatellite analysis on hepatocellular carcinoma. Mod Pathol 2002;15:1213–20. [DOI] [PubMed] [Google Scholar]
- 18.Koshikawa K, Nomoto S, Yamashita K, et al. Allelic imbalance at 1p36 in the pathogenesis of human hepatocellular carcinoma. Hepatogastroenterology 2004;51:186–91. [PubMed] [Google Scholar]


