Abstract
Background: The p73 gene is a p53 homologue that induces apoptosis and inhibits cell proliferation. N-terminal truncated isoforms of p73 (ΔNp73) act as dominant-negative inhibitors of wild-type p53 and TAp73 and result in tumour growth in nude mice.
Aims: To detect ΔNp73 expression in 24 benign prostatic hyperplasia samples, 33 prostate carcinomas, and five normal samples and to evaluate the relation between ΔNp73, TAp73 concentrations, and the clinicopathological characteristics of patients with prostate cancer.
Methods: TAp73 was determined by real time polymerase chain reaction (PCR); ΔNp73 and ΔN’p73 were assessed using reverse transcription PCR. western blotting was used to analyse protein expression. p53 mutation was determined by immunohistochemistry.
Results: A significant increase of ΔNp73 was seen in 20 of 33 carcinomas and 17 of 24 benign prostate hyperplasia tissues, but in none of the normal samples. None of the specimens expressed ΔN’p73. No significant relation was found between TAp73 expression and clinical parameters. The incidence of positive expression of ΔNp73 correlated with the Gleason score in prostate carcinomas. Cancer samples with wild-type p53 had significantly higher expression of ΔNp73 than p53 mutant cancers.
Conclusion: These data suggest a potential role for ΔNp73 in prostate cancer progression.
Keywords: ΔNp73, p73, p53, prostate cancer
Prostate cancer (PCa) is the most commonly diagnosed malignancy in the male population of the Western world and the incidence of PCa is rising rapidly in China.1 The molecular basis of PCa is still poorly understood. Moreover, the molecular mechanisms at the basis of the favourable outcome in patients with disseminated disease are not yet known, and understanding these mechanisms might have important implications.
The tumour suppressor gene p53 is the most commonly mutated gene in human cancer. The p53 gene encodes the p53 protein, which is a key regulator of the cell cycle, controlling the transition from G1 phase to S phase. Although alterations in the p53 gene are found in more than half of all human cancers, the role of p53 mutations in human prostate cancer is controversial. According to most studies, mutations in the p53 gene are rare in early, localised prostate cancer, whereas they are found in about 20–40% of advanced (metastatic and/or hormone refractory) prostate carcinomas.2,3
“p73 encodes a variety of protein isoforms through the use of alternative promoters at the 5′-end or differential splicing at the 3′-end”
p73 was recently identified as structurally and functionally homologous to tumour suppressor protein p53 from its sequence homology and gene organisation: a transactivation domain, a DNA binding domain, and an oligomerisation domain.4 As such, p73 can exert p53-like activities in various contexts, including transactivating p53 target genes, such as p21, MDM2, or BAX, leading to apoptosis and cell growth arrest.5 In contrast to the p53 gene, which encodes one major protein, p73 encodes a variety of protein isoforms through the use of alternative promoters at the 5′-end or differential splicing at the 3′-end.6 At the 5′-end, four different N-terminally truncated isoforms (ΔNp73, ΔN’p73, Ex2p73, and Ex2/3p73) have now been found in human cancers and/or cancer cells. Each of these isoforms lack all or most of the transactivation domain and therefore are collectively called ΔTAp73. ΔTAp73 products are generated either via alternative exon splicing of the P1 promoter transcript (to produce ΔN’p73, Ex2p73, and Ex2/3p73) or via alternative use of the P2 promoter in intron 3 (to produce ΔNp73).7,8 Of importance, the transcripts ΔNp73 and ΔN’p73 encode the same protein product. ΔNp73 acts as a potent transdominant inhibitor of wild-type p53 and the transactivation competent TAp73 and confers drug resistance to the wild-type p53 harbouring tumour cells.9
This is the first report on an association between expression of ΔNp73 (ΔN’p73) and TAp73 in patients with prostate carcinoma and benign prostate hyperplasia (BPH). Our study was designed to detect ΔNp73 using reverse transcription polymerase chain reaction (RT-PCR) and western blot analysis to evaluate the relation between ΔNp73 expression and the biological parameters of the patients.
MATERIAL AND METHODS
Cell culture
LNCaP, DU145, SP3031, 22Rv1, PC-3, and K-562 cells were obtained from the American Type Culture Collection (Rockville, Maryland, USA). They were cultured in RPMI 1640 (HyClone, Logan, Utah, USA) containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 µg/ml streptomycin in a 5% CO2 atmosphere at 37°C.
Patients and tissues
BPH samples were obtained from 24 patients after transurethral resection. Clinically localised prostate tumours were obtained from 33 patients undergoing radical prostatectomy. Staging was assessed after pathological examination of formalin fixed specimens on the basis of the 1997 TNM classification. Clinical and biological data from the patients are provided (table 1). Five normal prostate samples were obtained at necropsy, eight to 10 hours after death, from five men ages 20–38 years without a history of reproductive or endocrine disease.
Table 1.
Patients’ clinical and histological characteristics
| PCa (%) | BPH (%) | |
| Cases | 33 | 24 |
| Mean (SD) age in years | 64.7 (3.9) | 66.1 (4.4) |
| TNM | ||
| T1 | 2 (6.1) | |
| T2 | 14 (42.4) | |
| T3 | 17 (51.5) | |
| Gleason score | ||
| 2–4 | 7 (21.2) | |
| 5–7 | 15 (45.5) | |
| 8–10 | 11 (33.3) | |
| Mean (SD) PSA (ng/ml) | ||
| <4.0 | 3 (9.1) | 9 (37.5) |
| 4.0–10.0 | 6 (18.2) | 12 (50.0) |
| >10.0 | 24 (72.7) | 3 (12.5) |
BPH, benign prostate hyperplasia; PCa, prostate cancer; PSA, prostate specific antigen.
Real time PCR and RT-PCR
Total RNA was extracted from cells and tissues using the RNAeasy minikit (Qiagen, Hilden, Germany). Total RNA (2 μg) was reverse transcribed using Superscript II RT (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s protocol. Real time PCR was performed using the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, California, USA) with SYBR green for determination of TAp73 mRNA. Reactions were performed in a total volume of 20 µl, including 10 µl 2× SYBR Green PCR master mix (Applied Biosystems), 5 µl of each primer at a 5 µM concentration, and 1 µl of the previously reverse transcribed cDNA template. The cycling conditions were as follows: 10 minutes at 95°C, and 40 cycles of 15 seconds at 95°C followed by one minute at 60°C.
Calculations were performed using values of average cycle threshold (Ct) as the calibrator from normal samples. For each experimental sample, the Ct of the target, normalised to an endogenous reference and relative to the calibrator, is given by: 2−ΔΔCt.
Routine RT-PCR for detection of ΔNp73 and ΔN’p73 was carried out using HotStarTaq DNA polymerase (Qiagen). The authenticity of the RT-PCR products was verified by sequencing. Tumour samples were defined as ΔNp73 or ΔN’p73 positive if a PCR amplification band was detectable after 35 cycles of amplification. The primers are as follows: TAp73 sense, 5′- GCACCACGTTTGAGCACCTCT-3′ and antisense, 5′-GCA GATTGAACTGGGCCATGA-3′; ΔNp73 sense, 5′-CAAACGGCCCGCATGTTCCC-3′ and antisense, 5′-TGGTCCATGGTGCTGCTCAGC-3′; ΔN’p73 sense, 5′-TCGACCTT CCCCAGTCAAGC-3′ and antisense, 5′- TGGGACGAGGCATGGATCTG-3′; and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) sense; 5′-GAAGGTGAAGGTCGGAGTCA-3′ and antisense, 5′-GAAGATGGTGATGGGATTTC-3′. K-562 was used as the positive control for RT-PCR analysis of ΔNp73 and ΔN’p73.
Immunohistochemical detection of mutant p53
Tissue sections (5 µm thick) of fixed, paraffin wax embedded specimens were cut, mounted on poly-L-lysine slides (Superfrost Plus; VWR, Milan, Italy), and then dewaxed in Histoclear. Specimens were rehydrated by sequential washing in ethanol solutions. The sections were then immersed in 0.3% hydrogen peroxide in methanol for 20 minutes at room temperature to block endogenous peroxidase activity. After non-specific sites had been blocked with 5% normal goat serum in phosphate buffered saline (PBS) for one hour, the sections were incubated with anti-p53 (pAb240; Chemicon, Temecula, California, USA) monoclonal antibody overnight at 4°C. In subsequent steps, we used a Vectastain ABC kit (Vector, Burlingame, California, USA) and DAB (diaminobenzidine) as a chromogen. The sections were then counterstained with haematoxylin. In each immunohistochemical staining, we performed additional staining without primary antibody in parallel as a negative control. Expression was classified as negative if < 5% of tumour cells were positive and as positive if > 5% were positive. Positive p53 immunostaining has been reported to be related to p53 mutations that increase the half life of the p53 protein, leading to intracellular accumulation.10,11 Therefore, from now on we will refer to immunodetectable p53 as mutant p53.
Western blot analysis
Specimens were homogenised in Triton X-100 buffer with 500mM HEPES (pH.7.0), 150mM NaCl, 10% glycerol, 1mM EDTA, and 1.2% Triton X-100. The homogenate was centrifuged at 1000 ×g for 15 minutes to remove whole cells, nuclei, and mitochondria. The supernatant was used to measure the protein concentration. An equal amount of protein (30 µg) was loaded on to 10% polyacrylamide gels, followed by transfer to a nitrocellulose membrane sheet. The membranes were blocked with 5% skimmed milk in 0.01M PBS, pH 7.4. After incubation with primary antibodies, membranes were washed with PBS for 10 minutes three times and then incubated with horseradish peroxidase conjugated second antibody. Signals were detected with an ECL system (Amersham Pharmacia Biotech, St Louis, Missouri, USA).
The following antibodies were used for western blot: a monoclonal antibody against p73 (Ab-1; Calbiochem, Cambridge, Massachusetts, USA), a polyclonal antibody against ΔNp73 (sc-23429; Santa Cruz Biotechnology, Santa Cruz, California, USA), and a monoclonal anti-GAPDH antibody (Chemicon)
Statistical analysis
The association between ΔNp73 expression, TAp73, expression, and other prognostic factors and the differences in the distribution of the groups of patients were assessed by the χ2 test or Fisher’s exact test. All analyses ware two tailed and were considered significant when p < 0.05.
RESULTS
Expression of TAp73
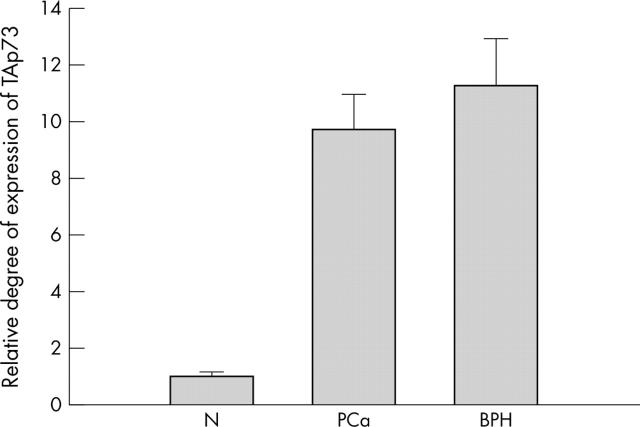
A change of threefold or greater was considered significant for TAp73 expression. The degree of expression was determined as the ratio between the TAp73 gene tested and the reference GAPDH gene to correct for variations in the amounts of RNA. Ratios were then normalised such that the mean ratio of the five normal prostate samples equalled 1.00. Overexpression of TAp73 was seen in 24 of 33 primary carcinomas, in three of five carcinoma cell lines (LNCaP, SP3031, and 22Rv1), and in 19 of 24 BPH specimens. The mean ratio of the TAp73 of the PCa and BPH samples was 9.65 and 11.21 times the mean ratio of the normals, respectively (p < 0.01) (fig 1).
Figure 1.
Quantification of TAp73 in prostate carcinoma (PCa), benign prostatic hyperplasia (BPH), and normal tissue (N). The relative expression levels were then normalised such that the mean ratio of the five normal prostate samples equalled 1.00. The mean ratio of TAp73 expression in the PCa and BPH samples was 9.65 and 11.21 times higher than the mean ratio of the normals, respectively.
Expression of ΔNp73
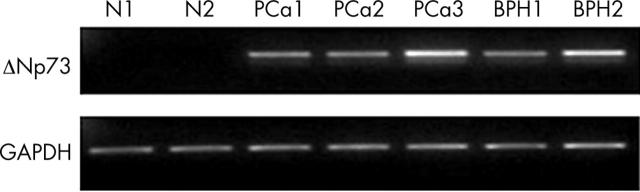
The normal tissues did not express detectable amounts of ΔNp73 transcripts, as determined by RT-PCR. Most of the tumours and BPH specimens showed increased expression of ΔNp73 (fig 2). A significant increase in ΔNp73 was seen in 20 of 33 tumours, 17 of 24 BPH samples, and two of five carcinoma cell lines (LNCaP and 22 Rv1). We were unable to detect ΔN’p73 in the samples. Nucleic acid sequencing revealed that the positive band represents the N-terminal truncated isoform ΔNp73. We failed to reveal a relation between TAp73 and ΔNp73 mRNA expression in PCa and BPH samples (tables 2, 3). Table 4 summarises the various clinicopathological characteristics of the patients with cancer in relation to ΔNp73 and TAp73 mRNA expression. Positive ΔNp73 expression correlated with tumour Gleason score (p = 0.037). There was no significant relation between ΔNp73 expression and age at surgery, TNM stage, or PSA serum concentration. There was also no association between TAp73 expression and these clinical parameters.
Figure 2.
Expression of ΔNp73 (256 bp) in representative patient samples analysed by reverse transcriptase polymerase chain reaction (RT-PCR). The expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (226 bp) was analysed as an internal control. Authenticity of the RT-PCR products was determined by sequencing. BPH, benign prostate hyperplasia; N, normal, PCa, prostate carcinoma.
Table 2.
Correlation between TAp73 expression and ΔNp73 in prostate carcinoma tissues
| ΔNp73 expression | p Value | ||
| Positive | Negative | ||
| TAp73 expression | |||
| Positive | 17 | 7 | 0.107 |
| Negative | 3 | 6 | |
Table 3.
Correlation between TAp73 expression and ΔNp73 in benign prostate hyperplasia tissues
| ΔNp73 expression | p Value | ||
| Positive | Negative | ||
| TAp73 expression | |||
| Positive | 15 | 4 | 0.126 |
| Negative | 2 | 3 | |
Table 4.
Relation between ΔNp73, TAp73, and clinicopathological characteristics in 33 patients with prostate carcinoma
| ΔNp73 expression | p Value | TAp73 expression | p Value | |||
| Positive | Negative | Positive | Negative | |||
| Age at diagnosis | ||||||
| <65 | 11 | 7 | 0.717 | 13 | 5 | 1 |
| ⩾65 | 9 | 6 | 11 | 4 | ||
| TNM | ||||||
| T1a–T2b | 8 | 8 | 0.296 | 10 | 6 | 0.259 |
| T3a–T3b | 12 | 5 | 14 | 3 | ||
| Gleason score | ||||||
| 2–6 | 6 | 9 | 0.037 | 9 | 6 | 0.239 |
| 7–10 | 14 | 4 | 15 | 3 | ||
| PSA (ng/ml) | ||||||
| <10.0 | 5 | 4 | 1 | 8 | 1 | 0.384 |
| ⩾10.0 | 15 | 9 | 16 | 8 | ||
PSA, prostate specific antigen.
Western blotting
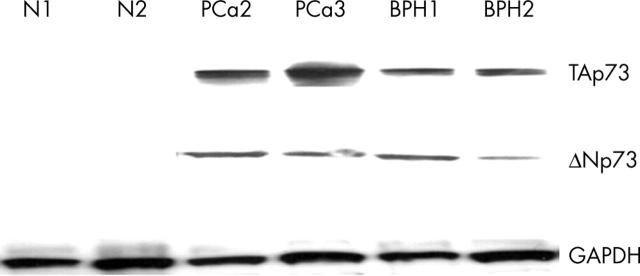
To verify whether our RT-PCR results reflected p73 protein expression, selected samples were analysed by western blotting using an antibody that recognises an epitope in the N-terminal transactivation domain of p73 and an antibody against ΔNp73, which recognises ΔNp73 only, with no crossreactivity with TAp73 isoforms. Both TAp73 and ΔNp73 expression were found in the tumour and BHP tissues, but not in normal tissues. As shown in fig 3, protein concentrations correlated with the RT-PCR data.
Figure 3.
Western blot analysis of representative patient samples. The tissue was lysed and 30 µg of cleared lysate was applied to a 10% acrylamide gel. TAp73 and ΔNp73 were shown to be present in the prostate carcinomas (PCa) and benign prostatic hyperplasia (BPH) samples, but not in the normal (N) tissues. GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Immunohistochemistry
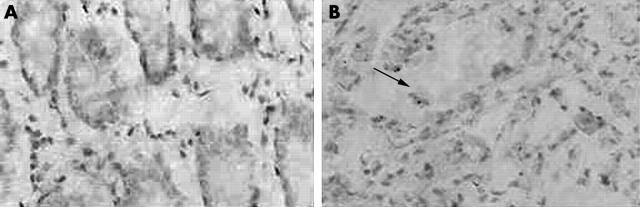
We determined the p53 mutation status in our tumour collection via immunohistochemistry. Ten of 33 tumours and one of 24 BPH specimens expressed mutant p53 (fig 4). Significantly higher expression of the ΔNp73 isoform was found in p53 wild-type tumours, compared with p53 mutant tumours (p = 0.026). In contrast, there was no difference in the prevalence of TAp73 upregulation between wild-type and mutant tumours (table 5).
Figure 4.
Representative examples of p53 immunohistochemical staining. (A) p53 expression in normal prostate tissue, (B) p53 expression in prostate carcinoma (arrow; original magnification, ×400).
Table 5.
Correlation between p53 status and p73 isoform expression in 33 prostate carcinomas
| p53 expression | p Value | ||
| Mutant | Wild-type | ||
| TAp73 expression | |||
| Positive | 6 | 18 | 0.400 |
| Negative | 4 | 5 | |
| ΔNp73 expression | |||
| Positive | 3 | 17 | 0.026 |
| Negative | 7 | 6 | |
DISCUSSION
We analysed the expression of two N-terminally truncated p73 isoforms in 33 prostate carcinomas and 24 BPHs of all histological types, and correlated the results with clinicopathological parameters. The most striking finding was the high prevalence of upregulation of ΔNp73 in prostate specimens (20 of 33 PCa and 17 of 24 BPH specimens), suggesting that deregulation of ΔNp73 is a crucial event in the pathogenesis of this tumour. ΔNp73 is derived from the P2 promoter. In contrast, none of the specimens showed upregulation of the P1 promoter derived ΔN’p73 transcript. We conclude that ΔNp73, rather than ΔN’p73, is the main contributor to total ΔNp73 upregulation in prostate cancers.
Increased expression of wild-type p73 in tumour samples compared with normal tissues has been reported frequently,12–15 and in some cases overexpression of p73 correlated with an advanced tumour stage or poor prognostic parameters.16,17 Consistent with our findings, increased TAp73 expression has been reported previously in prostate tumours compared with normal tissue.18 As shown as table 4, no significant differences were detected concerning median age, TNM stage, Gleason score, or PSA serum concentration, indicating that TAp73 expression cannot be considered as a prognostic marker. However, ΔNp73 expression was strongly associated with the Gleason score, suggesting a potential role of ΔNp73 transcripts in prostate carcinoma progression, because the Gleason score is considered one of the most important prognostic indicators for prostate cancer. Our results are also in agreement with two recent ΔNp73 studies,19,20 which both found significantly higher expression of ΔNp73 in advanced tumours.
“ΔNp73 expression was strongly associated with the Gleason score, suggesting a potential role of ΔNp73 transcripts in prostate carcinoma progression”
Most human tumours show deregulation of the E2F1 family of transcription factors through the loss of cyclin dependent kinase inhibitor INK4, overexpression of cyclin D, or loss of retinoblastoma protein. E2F1 directly regulates p73 by binding to E2F1 binding sites within the P1 promoter. Interestingly, we found that the P2 promoter derived ΔNp73 transcript was more abundant in benign prostate hyperplasia (17 of 24) than in PCa (20 of 33). In line with our observations, previous results showed that ΔNp73, the predominant isoform in benign naevi, was not upregulated significantly in melanoma tissue.21 This might be because the P2 promoter has a lower sensitivity for E2F1, as indicated by a previous study on Saos-2 cells,22 where there was a strong induction of P1 promoter derived transcripts after conditional activation of E2F1, whereas the P2 promoter regulated ΔNp73 transcript showed only a modest induction by E2F1. Another possible reason is that ΔNp73 expression is epigenetically regulated in prostate tissues. It was reported previously that methylation of the ΔNp73 5′-region in neuroblastoma was associated with the absence of expression.23 We speculate that differences in methylation of ΔNp73 contributes to this discrepancy. It is currently unclear why the P1 promoter derived ΔN’p73 transcript is hardly expressed in prostate neoplastic cells. One possible explanation might be that different splice factors are required for the generation of aberrantly spliced transcripts from the P1 promoter.
Take home messages.
Expression of the N-terminally truncated isoform of p73, ΔNp73, was high in 20 of 33 prostate carcinomas and 17 of 24 benign prostate hyperplasia tissue samples, but not in normal samples
Positive ΔNp73 expression correlated with the Gleason score in prostate carcinoma
These results suggest a potential role for ΔNp73 in prostate cancer progression
Another surprising result of our study is the strong correlation between p53 status and the upregulation of dominant-negative ΔNp73 in tumours. Considering the fact that oncogene induced upregulation of p73 expression causes apoptosis, sustained overexpression of p73 therefore requires inhibition of its inherent proapoptotic activity.24 This could be achieved by p53 mutants, which inhibit the putative tumour suppressor action of p73 in a dominant-negative fashion by generating defective hetero-oligomers with wild-type p73. However, reduction of the apoptotic potential of p73 by mutant p53 cannot account for all cases of p73 overexpression. In our study, no correlation between p53 mutations and p73 overexpression could be detected in prostate carcinoma. In this respect, the interaction between p53 and ΔNp73 might contribute to malignancy in the absence of physical damage to the p53 gene. p53 is capable of inducing ΔNp73, a protein that can block p53 action by competing for p53 DNA binding sites. p53 induces ΔNp73 by directly binding to its promoter.25 ΔNp73 counteracts p53 action on its own promoter, creating an additional feedback loop that finely tunes the entire system. In addition, a similar loop regulates TAp73 action, because ΔNp73 expression is also induced by TAp73, and ΔNp73 also regulates the function of the TA forms. This suggests that ΔNp73 plays an important role in regulating p53 function and that its overexpression could play a role in tumorigenesis by generating a functional block of p53. We reasoned that if ΔNp73 isoforms are indeed oncogenic inhibitors of p53 and TAp73 in vivo, their upregulation should occur preferentially in wild-type p53 tumours, and our results support this notion.
In conclusion, our study identified a novel biological feature of PCa and BPH. Additional laboratory studies are needed to elucidate the genetic mechanism underlying the frequent expression of ΔNp73 in PCa and BPH and its biological consequences on cell survival.
Acknowledgments
This work was supported in part by National Natural Science Foundation of China (No. 30300360) and the “Raising Star” plan from Shanghai Science and Technology Committee.
Abbreviations
ΔN, N truncated
BPH, benign prostate hyperplasia
Ct, cycle threshold
GAPDH, glyceraldehyde 3-phosphate dehydrogenase
PBS, phosphate buffered saline
PCa, prostate carcinoma
PCR, polymerase chain reaction
PSA, prostate specific antigen
RT, reverse transcriptase
REFERENCES
- 1.Gu F. Epidemiological survey of benign prostatic hyperplasia and prostatic cancer in China. Chin Med J (Engl) 2000;113:299–302. [PubMed] [Google Scholar]
- 2.Bookstein R, MacGrogan D, Hilsenbeck SG, et al. p53 is mutated in a subset of advanced-stage prostate cancers. Cancer Res 1993;53:3369–73. [PubMed] [Google Scholar]
- 3.Visakorpi T, Kallioniemi OP, Heikkinen A, et al. Small subgroup of aggressive, highly proliferative prostatic carcinomas defined by p53 accumulation. J Natl Cancer Inst 1992;84:883–7. [DOI] [PubMed] [Google Scholar]
- 4.Kaghad M, Bonnet H, Yang A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997;90:809–19. [DOI] [PubMed] [Google Scholar]
- 5.Jost CA, Marin MC, Kaelin WG Jr. p73 is a human p53-related protein that can induce apoptosis. Nature 1997;389:191–4. [DOI] [PubMed] [Google Scholar]
- 6.Yang A, Kaghad M, Caput D, et al. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet 2002;18:90–5. [DOI] [PubMed] [Google Scholar]
- 7.Ishimoto O, Kawahara C, Enjo K, et al. Possible oncogenic potential of deltaNp73: a newly identified isoform of human p73. Cancer Res 2002;62:636–41. [PubMed] [Google Scholar]
- 8.Kartasheva NN, Contente A, Lenz-Stoppler C, et al. p53 induces the expression of its antagonist p73 delta N, establishing an autoregulatory feedback loop. Oncogene 2002;21:4715–27. [DOI] [PubMed] [Google Scholar]
- 9.Zaika AI, Slade N, Erster SH, et al. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med 2002;196:765–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson P, Ow K, Yardley G, et al. Downregulation of KAI1 mRNA in localised prostate cancer and its bony metastases does not correlate with p53 overexpression. Prostate Cancer Prostatic Dis 2003;6:174–81. [DOI] [PubMed] [Google Scholar]
- 11.Mehta R, Kyshtoobayeva A, Kurosaki T, et al. Independent association of angiogenesis index with outcome in prostate cancer. Clin Cancer Res 2001;7:81–8. [PubMed] [Google Scholar]
- 12.Tannapfel A, Wasner M, Krause K, et al. Expression of p73 and its relation to histopathology and prognosis in hepatocellular carcinoma. J Natl Cancer Inst 1999;91:1154–8. [DOI] [PubMed] [Google Scholar]
- 13.Guan M, Peng HX, Yu B, et al. p73 Overexpression and angiogenesis in human colorectal carcinoma. Jpn J Clin Oncol 2003;33:215–20. [DOI] [PubMed] [Google Scholar]
- 14.Pan H, Liao SJ, Lai WY, et al. Overexpression but lack of mutation and methylation of p73 in hepatocellular carcinoma. Acta Oncol 2002;41:550–5. [DOI] [PubMed] [Google Scholar]
- 15.Sun XF. p73 overexpression is a prognostic factor in patients with colorectal adenocarcinoma. Clin Cancer Res 2002;8:165–70. [PubMed] [Google Scholar]
- 16.Nozaki M, Tada M, Kashiwazaki H, et al. p73 is not mutated in meningiomas as determined with a functional yeast assay but p73 expression increases with tumor grade. Brain Pathol 2001;11:296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novak U, Grob TJ, Baskaynak G, et al. Overexpression of the p73 gene is a novel finding in high-risk B-cell chronic lymphocytic leukemia. Ann Oncol 2001;12:981–6. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi H, Ichimiya S, Nimura Y, et al. Mutation, allelotyping, and transcription analyses of the p73 gene in prostatic carcinoma. Cancer Res 1998;58:2076–7. [PubMed] [Google Scholar]
- 19.Uramoto H, Sugio K, Oyama T, et al. Expression of deltaNp73 predicts poor prognosis in lung cancer. Clin Cancer Res 2004;10:6905–11. [DOI] [PubMed] [Google Scholar]
- 20.Ugur H, Sayan AE, Ozdamar SO, et al. Expression of TAP73 and deltaNP73 in malignant gliomas. Oncol Rep 2004;11:1337–41. [PubMed] [Google Scholar]
- 21.Tuve S, Wagner SN, Schittek B, et al. Alterations of deltaTA-p73 splice transcripts during melanoma development and progression. Int J Cancer 2004;108:162–6. [DOI] [PubMed] [Google Scholar]
- 22.Stiewe T, Zimmermann S, Frilling A, et al. Transactivation-deficient deltaTA-p73 acts as an oncogene. Cancer Res 2002;62:3598–602. [PubMed] [Google Scholar]
- 23.Casciano I, Banelli B, Croce M, et al. Role of methylation in the control of deltaNp73 expression in neuroblastoma. Cell Death Differ 2002;9:343–5. [DOI] [PubMed] [Google Scholar]
- 24.Stiewe T, Putzer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Gene 2000;26:464–9. [DOI] [PubMed] [Google Scholar]
- 25.Grob TJ, Novak U, Maisse C, et al. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ 2001;8:1213–23. [DOI] [PubMed] [Google Scholar]