Abstract
Background: Extrahepatic biliary stenosis (EBS) has malignant and benign causes. Patients with EBS are at risk of having or developing malignancy. Accurate diagnostic tests for early detection and surveillance are needed. The sensitivity of biliary cytology for malignancy is low. K-ras mutation analysis on brush cytology is a valuable adjunct, but specificity is low. A quantitative test for K-ras mutations has been developed: the amplification refractory mutation system (ARMS).
Aim: To assess the test characteristics and additional value of ARMS in diagnosing the cause of EBS.
Methods: Brush samples from endoscopic retrograde cholangiopancreatography were collected from 312 patients with EBS. K-ras mutation analysis was performed using ARMS—allele specific amplification was coupled with real time fluorescent detection of PCR products. Results were compared with conventional cytology and K-ras mutation analysis using allele specific oligonucleotide (ASO) hybridisation, and evaluated in view of the final diagnosis.
Results: The test characteristics of ARMS and ASO largely agreed. Sensitivity for detecting malignancy was 49% and 42%, specificity 93% and 88%, and positive predictive value (PPV) 96% and 91%, respectively. The sensitivity of ARMS and cytology combined was 71%, and PPV was 93%. The specificity of ARMS could be increased to 100% by setting limits for the false positives, but reduced sensitivity from 49% to 43%.
Conclusions: ARMS can be considered supplementary to conventional cytology, and comparable to ASO in diagnosing malignant EBS. A specificity of 100% can be achieved with ARMS, which should be considered in the surveillance of patients at risk for pancreatic cancer.
Keywords: K-ras, amplification refractory mutation system, allele specific oligonucleotide, brush cytology, extrahepatic biliary stenosis
Differentiating between malignant and benign causes of extrahepatic biliary stenosis (EBS) is often difficult, but very important.1–5 Patients with EBS are at risk of having or developing pancreatic cancer. Others at risk are family members of patients with pancreatic cancer, a fact that has been recognised for decades. Both groups have an increased risk of developing pancreatic cancer, and the application of a screening test for early detection would be very helpful. Because the incidence of pancreatic cancer in the general population is relatively low, screening tests should be limited to the abovementioned groups with increased risk.
“The localisation of most K-ras mutations to the single codon 12 makes them relatively easy to detect”
Although the specificity of brush cytology for a malignant cause of EBS obtained during endoscopic retrograde cholangiopancreatography (ERCP) is high,6–8 its sensitivity is relatively low.9 Previously, we showed that the addition of K-ras mutation analysis using allele specific oligonucleotide (ASO) hybridisation to brush cytology improved the sensitivity of diagnosing a malignancy.7 Brush sampling during ERCP has a high probability of yielding sufficient cells for DNA analysis, and these cells may contain mutations that originated in malignant cells preferentially shed from pancreatic ducts. Using these sample, it may also be possible to detect cells originating from other malignant causes of EBS, such as distal bile duct carcinoma, in which K-ras mutations have also been reported.10 The 89% specificity of K-ras mutation analysis based on data from 312 consecutive patients with EBS in our study is one of the highest reported. Nevertheless, a diagnostic test with a specificity of 89% remains suboptimal, especially considering the major therapeutic consequences of a malignant diagnosis.7,11
Despite its limitations, we think that K-ras is a promising marker in the diagnosis of EBS. The cause of malignant EBS is mostly pancreatic head carcinoma, which has the highest incidence of RAS mutations in human tumours identified to date.12–14 Furthermore, the localisation of most K-ras mutations to the single codon 12 makes them relatively easy to detect.15,16 Depending on the technique used, the reported frequencies of codon 12 mutations range from 20% to 100%, and occur as early events in tumour progression.12,13,17,18
Recently, a novel quantitative real time assay for K-ras mutations has been developed: the amplification refractory mutation system (ARMS) assay.19,20 The real time quantitative approach of this assay, with its sensitive detection and mutant sequence quantification, allows the determination of the true detection limit in any clinical application.20 It routinely provides quantitative data relating to the number of K-ras mutations in positive samples, allowing a threshold to be set, above which the specificity will be 100%.
A quantitative real time assay might be a valuable adjunct to early detection in patients with EBS, and also to surveillance strategies in family members of patients with pancreatic cancer. The genetic transmission of pancreatic cancer in hereditary familial syndromes and those patients at high risk are increasingly being better defined.21 There is a need for a molecular diagnostic test for members of families with certain pancreatic cancer syndromes, which could translate recent molecular genetic discoveries into improved surveillance measures. The ARMS assay for K-ras mutation analysis might be such a test.
The aim of our study was to assess the value of the quantitative ARMS assay for K-ras mutations compared with conventional cytopathology and the established enriched polymerase chain reaction (PCR)–ASO assay for the diagnosis of malignancy in patients with EBS in a large series of consecutive patients with complete follow up.
MATERIALS AND METHODS
Patients
Two earlier studies have been published on the same patient cohort.7,11 The study population consisted of 312 consecutive patients who underwent ERCP with endobiliary brush cytology for the evaluation of EBS at the Academic Medical Center in Amsterdam, the Netherlands from January 1993 to February 1996. The mean age of the 312 patients was 63 (range, 19–99) years and the male to female ratio was 173 : 139. The follow up was updated and four additional final diagnoses were encountered that were previously unspecified. For this study, a final diagnosis could be made in 298 patients, 223 (75%) of whom had malignant and 75 (25%) benign stenosis. Table 1 shows the spectrum of the different aetiologies of EBS.
Table 1.
Spectrum of the different causes of extrahepatic biliary stenosis in 298 patients with a final diagnosis
| Aetiology | Number of patients (N = 298) |
| Malignant stenosis | 223 (75%) |
| Pancreatic carcinoma | 98 |
| Bile duct carcinoma | 73 |
| Gall bladder carcinoma | 7 |
| Ampullary carcinoma | 8 |
| Lymph node metastasis | 11 |
| Lymphoma | 1 |
| Unspecified | 25 |
| Benign stenosis | 75 (25%) |
| Inflammatory | |
| Chronic pancreatitis | 27 |
| Cholelithiasis | 3 |
| Mirizzi syndrome | 1 |
| Primary sclerosing cholangitis | 26 |
| Postsurgical | 13 |
| Unspecified | 5 |
Samples and DNA isolation
The methods for collecting the required samples and isolating the DNA have been described previously.7 In summary, after brushing of the EBS four cytology smears from each patient underwent conventional Giemsa and Papanicolaou staining and were independently evaluated by an experienced cytopathologist. The following diagnostic categories were used: positive for carcinoma, negative for carcinoma, suspicious for carcinoma, and material insufficient or not suitable for diagnosis. The remaining brush specimen was suspended in 10 ml of DNA buffer, fixed with 10 ml 100% ethanol, and stored at 4°C for subsequent K-ras mutation analysis. The archival tissue blocks, available from 71 patients with malignant EBS and 10 patients with benign EBS, were analysed for K-ras mutations.
A 1 ml aliquot of each brush cytology suspension was used for DNA isolation. Careful microdissection of the tissue blocks was performed to ascertain a sample where at least 50% of the cells comprised the tissue of interest. DNA was extracted as described previously.22
K-ras mutation analyses
Two different methods were used for K-ras mutation analysis: the PCR–ASO hybridisation based assay and the novel ARMS allele specific amplification assay.
The protocol for the first method has been described and validated previously.7,22,23 With this assay, DNA is subjected to PCR amplification using primers around codon 12. Digestion of the PCR products with a restriction enzyme is followed by a second round of amplification, which yields a PCR product enriched for K-ras codon 12 mutations. The resulting DNA fragments are denatured and dot blotted on to nylon membranes and subjected to allele specific oligonucleotide hybridisation with radioactively labelled probes specific for each possible K-ras codon 12 mutation, followed by autoradiography. Controls for positive and negative outcomes, contamination, and specific and non-specific hybridisation were used. Both enriched and non-enriched PCR products were dot blotted next to each other to check the digestion and mutant enrichment.
The second method was based on ARMS allele specific amplification for mutant K-ras sequence discrimination. This was undertaken using the ABI 7700 machine (PE Applied Biosystems, Foster City, California, USA) to detect amplification products by fluorescence in real time. This assay has been described in detail.19,20 In summary, 5 ml of each of the 11 brush cytology DNA samples was added to each of seven ARMS reactions and a control reaction for DNA in a 96 tube format. An additional control reaction without DNA for each ARMS mix was included to detect possible contamination. The control reaction amplified all K-ras exon 1 sequences, irrespective of mutational status, to measure the total amount of DNA in each sample. Reactions were thermocycled in the ABI 7700 cycler and the relative fluorescence was measured after each cycle. The point at which it exceeded a threshold baseline signal was called the threshold cycle. Threshold cycle values from the control and ARMS reactions were plotted against statistically validated data obtained using wild-type K-ras exon 1. These data were used to establish the background signal resulting from wild-type K-ras exon 1 over a wide dynamic range of starting DNA concentrations (100 fold). The dynamic range of starting DNA concentrations (6–600 ng DNA) was chosen in relation to the yield of DNA typically obtained from 5 ml of DNA solution extracted from all clinical samples (tissue and cell suspensions in bodily fluids) entering our laboratory. Any clinical sample giving a signal above background in an ARMS reaction had a < 1% probability of containing wild-type sequence alone, and was therefore interpreted as positive for that mutation. A similar collection of data was obtained using each of the seven mutant K-ras sequences over the same concentration range. In this way, the amount of K-ras mutant sequence in each positive sample could be calculated as a proportion of the total amount of K-ras exon 1 in that particular sample.
Definitions of test characteristics
Sensitivity was defined as the percentage of patients with malignancy and positive test results. Specificity was defined as the percentage of patients without malignancy and negative test results. Positive predictive value was defined as the percentage of patients with positive test results who had a malignancy. Negative predictive value was defined as the percentage of patients with negative test results who did not have a malignancy.
RESULTS
Eighty one of 223 (36%) patients with malignant EBS were found to be positive by cytology alone. By combining suspicious cytology results with positive cytology results, 111 of 223 (49%) malignancies could be detected. Ninety four of 223 (42%) patients with malignant EBS were detected by K-ras analysis using the enriched PCR–ASO assay alone, and 109 (49%) were detected by ARMS alone. Table 2 summarises the test results. When the results from the ARMS assay and cytology were combined, 129 of 223 (58%) and 159 of 223 (71%) confirmed malignancies were detected, depending on whether the cytology results were restricted to the positive samples or included the suspicious samples.
Table 2.
Distribution of ERCP cytology samples based on diagnostic procedure results and final diagnosis of malignancy*
| Diagnostic procedure | Diagnostic result | Malignancy present | Malignancy absent |
| Cytology | Positive | 81/223 (36%) | 2/75 (3%) |
| Negative | 105/223 (47%) | 67/75 (89%) | |
| Suspicious | 30/223 (13%) | 5/75 (7%) | |
| Insufficient material | 7/223 (3%) | 1/75 (1%) | |
| ASO | Positive | 94/223 (42%) | 9/75 (12%) |
| Negative | 129/223 (57%) | 66/75 (88%) | |
| ARMS | Positive | 109/223 (49%) | 5/75 (7%) |
| Negative | 114/223 (51%) | 70/75 (93%) |
*The final diagnosis of malignancy was established by histological and/or clinical findings (symptomatology, imaging studies, and course of the disease).
ARMS, amplification refractory mutation system; ASO, allele specific oligonucleotide; ERCP, endoscopic retrograde cholangiopancreatography.
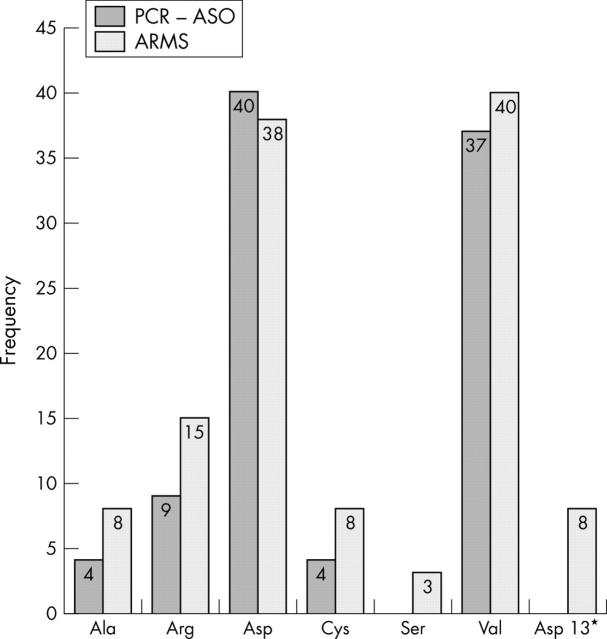
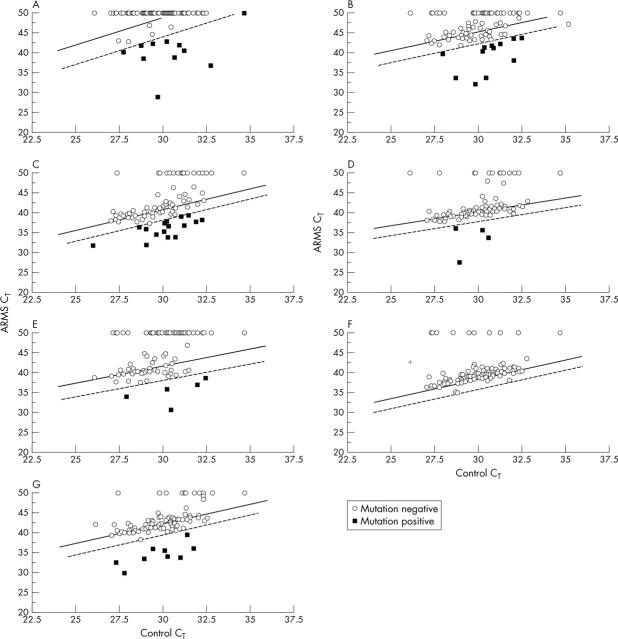
The results of the two different methods (the enriched PCR–ASO assay and the ARMS assay) were largely in agreement. Figure 1 shows the frequency of occurrence of each mutation detected by the two assays. The most frequently observed mutations in both assays resulted in codon 12 Gly → Asp and Gly → Val amino acid substitutions. No codon 12 Gly → Ser mutations were detected by enriched PCR–ASO. An additional test for the codon 13 Gly → Asp mutation in the ARMS assay detected eight mutations. There was no equivalent test in the enriched PCR–ASO assay and a zero was recorded for this mutation. Figure 2 shows the ARMS assay results for K-ras point mutations in 100 ERCP samples. Representation by this means has been described previously in lung cancer.20 Most samples in each test were negative for K-ras mutations. The data points appear above the 98% confidence interval established using wild-type K-ras exon 1, or fail to give ARMS products during cycling and are recorded as 50 cycles. Samples appearing below the lower limit of the 98% confidence interval have a < 1% chance of containing the wild-type sequence alone and are therefore recorded as positive. Duplicate analysis using the same DNA samples confirmed the positive samples (data not shown).
Figure 1.
Distribution of K-ras mutations detected by enriched PCR–ASO and by real time ARMS allele specific amplification. The bars represent the frequency of appearance of each mutation in the study. *There was no enriched PCR–ASO assay for the codon 13 Gly → Asp mutation and a zero result is displayed. ARMS, amplification refractory mutation system; ASO, allele specific oligonucleotide; PCR, polymerase chain reaction.
Figure 2.
Real time ARMS analysis of 100 ERCP brush cytology samples for K-ras mutations. The appearance of products in the ARMS and control reactions (CT) for each cytology sample plotted against data obtained from wild-type control DNA. ERCP samples were analysed in each of seven K-ras tests: (A) codon 12 GGT → GCT (Gly → Ala); (B) codon 12 GGT → CGT (Gly → Arg); (C) codon 12 GGT → GAT (Gly → Asp); (D) codon 13 GGC → GAC (Gly → Asp); (E) codon 12 GGT → TGT (Gly → Cys); (F) codon 12 GGT → AGT (Gly → Ser); and (G) codon 12 GGT → GTT (Gly → Val). The two regression lines represent mean Yi (solid line) and the 99% confidence limit for Yi data (dotted line). ARMS, amplification refractory mutation system; CT, threshold cycle; ERCP, endoscopic retrograde cholangiopancreatography.
All three diagnostic methods gave positive results in cases of confirmed non-malignancy. False positivity was 7% (five of 75) for the ARMS assay, 12% (nine of 75) for the PCR–ASO assay, and 3% (two of 75) to 7% (five of 75) for conventional cytology (table 2). Table 3 summarises the results of the 15 samples that had a false positive diagnostic test. Because the results of the ARMS assay were quantitative, cutoff limits were set for each of the four mutations found. These were 12.5%, 0.95%, 1.25%, and 0.24% for the codon 12 Gly → Ala, Gly → Asp, Gly → Cys, and Gly → Val tests, respectively. This means that a specificity of 100% was established for the diagnosis of malignancy using the adjusted ARMS assay. This reduced the sensitivity of the assay from 49% to 43%, because mutations were detected at equivalent or lower levels in samples from patients with clinically confirmed malignancy. Table 4 summarises the sensitivity, specificity, positive predictive value, and negative predictive value for all of the methods used. The main problem with all of the approaches used in isolation was that their negative predictive values were low and they did not detect enough malignancies. However, when ARMS and cytology were combined they were able to detect 159 of 223 (71%) malignancies with a positive predictive value of 93%. Individually, they detected 109 of 223 (49%) and 111 of 223 (50%) malignancies, with positive predictive values of 96% and 94%, respectively.
Table 3.
Summary of results for the 15 non-malignant* samples that gave positive results by cytology, PCR–ASO, or ARMS
| Patient’s sex/age (years) | Cytology positive | Cytology suspicious | ASO positive | ARMS positive* |
| Female/31 | – | – | 12 Asp | – |
| Male/60 | – | – | 12 Asp | – |
| Male/43 | – | – | Ala | Ala (9%) |
| Male/46 | – | + | – | – |
| Female/85 | – | – | Cys | – |
| Female/44 | – | + | – | – |
| Female/54 | – | – | – | Cys (1.25%) |
| Male/72 | – | + | – | – |
| Male/41 | – | – | 12 Asp | – |
| Male/38 | – | – | Ala | Ala (12.5%) |
| Male/44 | – | + | – | – |
| Female/60 | – | – | 12 Asp | 12 Asp (0.95%) |
| Male/55 | + | – | Val | – |
| Male/51 | – | + | – | – |
| Female/40 | + | – | 12 Asp | Val (0.24%) |
*Based on a final diagnosis of malignancy, which was established by histological and/or clinical findings (symptomatology, imaging studies, and course of the disease).
ARMS, amplification refractory mutation system; ASO, allele specific oligonucleotide; PCR, polymerase chain reaction.
Table 4.
Summary of the diagnostic specificity and sensitivity of cytology, PCR–ASO, and ARMS in relation to malignancy
| Diagnostic test result | Sensitivity | Specificity | PPV | NPV |
| Cytology (positive and suspicious) | 49% | 91% | 94% | 35% |
| Cytology (positive) | 36% | 97% | 98% | 39% |
| PCR–ASO | 42% | 88% | 91% | 34% |
| ARMS | 49% | 93% | 96% | 38% |
| ARMS (adjusted*) | 43% | 100% | 100% | 37% |
| Cytology (positive) + ARMS | 60% | 92% | 96% | 44% |
| Cytology (positive and suspicious) + ARMS | 71% | 84% | 93% | 53% |
*Adjusted ARMS results based on quantities of K-ras mutant sequence in confirmed non-malignant samples.
ARMS, amplification refractory mutation system; ASO, allele specific oligonucleotide; NPV, negative predictive value; PCR, polymerase chain reaction; PPV, positive predictive value.
DISCUSSION
Our study shows that the novel quantitative ARMS assay for K-ras mutation analysis is a valuable adjunct to conventional cytology and the non-quantitative PCR–ASO assay in diagnosing EBS. By setting cutoff limits, the ARMS assay was 100% specific and 43% sensitive in diagnosing a malignant cause in 312 consecutive endobiliary brush samples obtained from patients with EBS. Furthermore, a diagnostic combination of conventional cytology and the ARMS assay was able to predict a malignant cause in 93% of patients.
Cytology is highly specific in diagnosing the cause of EBS, but its sensitivity is routinely low.9,24 Although it has been suggested that repeated brushings increase the sensitivity,25 cytology alone is not sufficient to differentiate between malignant and benign EBS. Our study population comprised patients at risk of having or developing malignancy, mostly pancreatic or distal bile duct carcinoma.26–28 At present, the early detection of pancreatic cancer is the only realistic option for cure. Although screening for pancreatic cancer in the general population is not justified at the moment, there is certainly a need for accurate diagnostic tests in families with genetic disorders known to predispose to pancreatic cancer.21 Sensitive molecular assays offer the chance to improve test characteristics when used as an addition to cytology. Obviously, this mainly depends on the prevalence of the candidate marker in the disease and the technique used for the detection of molecular changes.
K-ras mutations are highly prevalent in pancreatic carcinoma13 and common in bile duct carcinoma, the two main malignant causes of EBS.26–28 K-ras encodes a protein located on the inner side of the plasma membrane, which has intrinsic GTPase activity. A mutation in the K-ras gene at codon 12 or 13 results in inappropriate growth signalling.29 Several studies have investigated the clinical usefulness of K-ras mutation analysis in the diagnosis and treatment of EBS. Reported rates of K-ras mutations and outcomes of the numerous studies vary widely. For example, Ponsioen et al found no additional value of K-ras mutation analysis in discriminating between benign and malignant strictures in patients with primary sclerosing cholangitis.30 However, we showed previously that K-ras codon 12 mutation analysis was a useful supplement to light microscopic evaluation of ERCP brush cytology specimens for the diagnosis of malignant EBS.7,11
“Although screening for pancreatic cancer in the general population is not justified at the moment, there is certainly a need for accurate diagnostic tests in families with genetic disorders known to predispose to pancreatic cancer”
Variations in reported diagnostic K-ras mutations can be attributed to the nature of the clinical material being investigated and the sensitivity and specificity of the assays used. In a previous study, we used fine needle aspiration of pancreatic or duodenal juice to show that a molecular panel including the K-ras, p53, and DPC4 (MAD4) genes can supplement traditional cytological diagnosis.31 Fine needle aspiration has the potential advantage of enriching for malignant cells.32 When secretin is administered as part of an exocrine function test before sampling, K-ras mutations can be seen in juice samples obtained from patients with benign disease.33,34 In our current study, we used brush samples obtained during ERCP, performed routinely in patients with EBS at our institution, as an indirect sampling method to reduce the chance of detecting cells from benign tissue.
Unfortunately, mutations in the K-ras oncogene also occur in non-malignant pancreatic tissue.18,35 K-ras gene mutations have been found in premalignant disease states such as pancreatic intraepithelial neoplasia lesions and chronic pancreatitis.18 In the recently developed progression model for pancreatic ductal adenocarcinoma, mutations in the K-ras oncogene seem to be an early event in the series of architectural and cytological changes.36–38 In a previous study on the same 312 consecutive patients, we performed a longterm follow up of the eight patients with a K-ras mutation detected in brushings of clinically benign EBS.11 After a median follow up of 65 months all eight were considered to be confirmed false positives. Few publications exist on patients who develop pancreatic cancer after an interval of more than 65 months.39,40 Although false positive results were infrequent, and in theory these eight patients could still develop malignancy, a diagnostic test with a specificity of 89% remains suboptimal.
The novel real time ARMS assay used in our current study provided quantitative data, allowing the determination of the true detection limit, above which the specificity was 100%.20 It is a convenient and homogeneous method, which facilitates high throughput sample analysis for a range of clinical materials. The enriched PCR–ASO assay requires separate amplification steps and will always carry the risk of PCR contamination. In contrast, the ARMS assay is a closed tube assay and the amplification products can be disposed of with a vastly reduced risk of contamination. The additional data provided by this sensitive technique used in conjunction with cytology increased the detection of clinically confirmed malignancy from 111 of 223 (50%) to 159 of 223 (71%). The 100% specificity in the 298 ERCP samples analysed by ARMS alone was based on the appearance of false positives in the clinically confirmed non-malignant cases. The occurrence of mutations in subsequent samples taken from patients with a clinical diagnosis of non-malignant stenosis will redefine the cutoff limits depending on the numbers of mutations. This raises questions about the importance of mutations detected at equivalent or lower levels in patients with confirmed malignancy. However, the aim of any diagnostic assay is to achieve the highest specificity when distinguishing between two clinical scenarios, while maintaining a reasonable sensitivity.
Knowledge about well defined high risk groups that might benefit from sensible surveillance strategies is rapidly increasing.41 Particularly noteworthy in this regard are the hereditary syndromes that include pancreatic cancer.41,42 A genetic marker of sufficiently high sensitivity and specificity is required to determine those unaffected relatives in the direct cancer prone lineage who are at increased risk of pancreatic cancer.43 The ARMS assay could be a valuable adjunct in this respect, because it can be set to a specificity of 100%. Members of families prone to pancreatic cancer are at high risk for the development of pancreatic cancer and therefore ideal candidates for surveillance.44 We believe that a symptomatic kindred of patients with a mutation known to be associated with familial susceptibility warrants an aggressive approach that incorporates our growing knowledge of the genetics of pancreatic cancer. The goal for the surveillance of patients with familial pancreatic cancer is to diagnose the malignancy in the dysplasia or carcinoma in situ stage, before the development of invasive cancer. Some authors even advise that a complete pancreaticoduodenectomy should be performed in these patients.45 It has already been shown that thorough screening of patients with a family history of pancreatic cancer is feasible.46 In the near future, other molecular markers of pancreatic cancer, recently discovered by global gene expression technology,47 could form a suitable diagnostic panel along with K-ras mutation analysis using the ARMS assay on brush cytology.
Take home messages.
The amplification refractory mutation system (ARMS) for K-ras mutation had a slightly higher sensitivity, specificity, and positive predictive value compared with the polymerase chain reaction–allele specific oligonucleotide assay in diagnosing patients with malignant extrahepatic biliary stenosis (EBS)
ARMS would be a useful supplement to conventional cytology in diagnosing patients with malignant EBS
A specificity of 100% can be achieved with ARMS, and this test should be considered in the surveillance of patients at risk for pancreatic cancer
In conclusion, our study shows that the novel, real time ARMS assay for K-ras mutations could be a useful supplement to conventional cytology and the non-quantitative PCR–ASO assay in diagnosing patients with malignant EBS. The quantitative nature of the ARMS assay makes it possible to diagnose malignant stenosis with a specificity of 100%.
Abbreviations
ARMS, amplification refractory mutation system
ASO, allele specific oligonucleotide
EBS, extrahepatic biliary stenosis
ERCP, endoscopic retrograde cholangiopancreatography
PCR, polymerase chain reaction
REFERENCES
- 1.Warshaw AL, Fernandez-del Castillo C. Pancreatic carcinoma. N Engl J Med 1992;326:455–65. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch-Ginsberg C. Detection of minimal residual disease: relevance for diagnosis and treatment of human malignancies. Annu Rev Med 1998;49:111–22. [DOI] [PubMed] [Google Scholar]
- 3.Leichman CG. Thymidylate synthase as a predictor of response. Oncology (Huntingt) 1998;12:43–7. [PubMed] [Google Scholar]
- 4.Findlay MP, Cunningham D, Morgan G, et al. Lack of correlation between thymidylate synthase levels in primary colorectal tumours and subsequent response to chemotherapy. Br J Cancer 1997;75:903–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson BR. Progress in determining the nature of bile duct strictures. Gut 1993;34:725–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JG, Leung JW, Baillie J, et al. Benign, dysplastic, or malignant—making sense of endoscopic bile duct brush cytology: results in 149 consecutive patients. Am J Gastroenterol 1995;90:722–6. [PubMed] [Google Scholar]
- 7.Sturm PD, Rauws EA, Hruban RH, et al. Clinical value of K-ras codon 12 analysis and endobiliary brush cytology for the diagnosis of malignant extrahepatic bile duct stenosis. Clin Cancer Res 1999;5:629–35. [PubMed] [Google Scholar]
- 8.Bardales RH, Stanley MW, Simpson DD, et al. Diagnostic value of brush cytology in the diagnosis of duodenal, biliary, and ampullary neoplasms. Am J Clin Pathol 1998;109:540–8. [DOI] [PubMed] [Google Scholar]
- 9.Ponchon T, Gagnon P, Berger F, et al. Value of endobiliary brush cytology and biopsies for the diagnosis of malignant bile duct stenosis: results of a prospective study. Gastrointest Endosc 1995;42:565–72. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe M, Asaka M, Tanaka J, et al. Point mutation of K-ras gene codon 12 in biliary tract tumors. Gastroenterology 1994;107:1147–53. [DOI] [PubMed] [Google Scholar]
- 11.van Heek NT, Rauws EA, Caspers E, et al. Long-term follow-up of patients with a clinically benign extrahepatic biliary stenosis and K-ras mutation in endobiliary brush cytology. Gastrointest Endosc 2002;55:883–8. [DOI] [PubMed] [Google Scholar]
- 12.Schneider G, Schmid RM. Genetic alterations in pancreatic carcinoma. Mol Cancer 2003;2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almoguera C, Shibata D, Forrester K, et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 1988;53:549–54. [DOI] [PubMed] [Google Scholar]
- 14.Sohn TA, Yeo CJ, Cameron JL, et al. Resected adenocarcinoma of the pancreas—616 patients: results, outcomes, and prognostic indicators. J Gastrointest Surg 2000;4:567–79. [DOI] [PubMed] [Google Scholar]
- 15.Sturm PD, Hruban RH, Ramsoekh TB, et al. The potential diagnostic use of K-ras codon 12 and p53 alterations in brush cytology from the pancreatic head region. J Pathol 1998;186:247–53. [DOI] [PubMed] [Google Scholar]
- 16.Wilentz RE, Chung CH, Sturm PD, et al. K-ras mutations in the duodenal fluid of patients with pancreatic carcinoma. Cancer 1998;82:96–103. [DOI] [PubMed] [Google Scholar]
- 17.Hruban RH, van Mansfeld AD, Offerhaus GJ, et al. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol 1993;143:545–54. [PMC free article] [PubMed] [Google Scholar]
- 18.Caldas C, Hahn SA, Hruban RH, et al. Detection of K-ras mutations in the stool of patients with pancreatic adenocarcinoma and pancreatic ductal hyperplasia. Cancer Res 1994;54:3568–73. [PubMed] [Google Scholar]
- 19.Fox JC, England J, White P, et al. The detection of K-ras mutations in colorectal cancer using the amplification-refractory mutation system. Br J Cancer 1998;77:1267–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clayton SJ, Scott FM, Walker J, et al. K-ras point mutation detection in lung cancer: comparison of two approaches to somatic mutation detection using ARMS allele-specific amplification. Clin Chem 2000;46:1929–38. [PubMed] [Google Scholar]
- 21.Klein AP, Beaty TH, Bailey-Wilson JE, et al. Evidence for a major gene influencing risk of pancreatic cancer. Genet Epidemiol 2002;23:133–49. [DOI] [PubMed] [Google Scholar]
- 22.Chung CH, Wilentz RE, Polak MM, et al. Clinical significance of K-ras oncogene activation in ampullary neoplasms. J Clin Pathol 1996;49:460–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hruban RH, Sturm PD, Slebos RJ, et al. Can K-ras codon 12 mutations be used to distinguish benign bile duct proliferations from metastases in the liver? A molecular analysis of 101 liver lesions from 93 patients. Am J Pathol 1997;151:943–9. [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrari AP Jr, Lichtenstein DR, Slivka A, et al. Brush cytology during ERCP for the diagnosis of biliary and pancreatic malignancies. Gastrointest Endosc 1994;40:140–5. [DOI] [PubMed] [Google Scholar]
- 25.Rabinovitz M, Zajko AB, Hassanein T, et al. Diagnostic value of brush cytology in the diagnosis of bile duct carcinoma: a study in 65 patients with bile duct strictures. Hepatology 1990;12:747–52. [DOI] [PubMed] [Google Scholar]
- 26.Motojima K, Tsunoda T, Kanematsu T, et al. Distinguishing pancreatic carcinoma from other periampullary carcinomas by analysis of mutations in the Kirsten-ras oncogene. Ann Surg 1991;214:657–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rijken AM, van Gulik TM, Polak MM, et al. Diagnostic and prognostic value of incidence of K-ras codon 12 mutations in resected distal bile duct carcinoma. J Surg Oncol 1998;68:187–92. [DOI] [PubMed] [Google Scholar]
- 28.Suto T, Habano W, Sugai T, et al. Aberrations of the K-ras, p53, and APC genes in extrahepatic bile duct cancer. J Surg Oncol 2000;73:158–63. [DOI] [PubMed] [Google Scholar]
- 29.Rashid A. Cellular and molecular biology of biliary tract cancers. Surg Oncol Clin N Am 2002;11:995–1009. [DOI] [PubMed] [Google Scholar]
- 30.Ponsioen CY, Vrouenraets SM, van Milligen de Wit AW, et al. Value of brush cytology for dominant strictures in primary sclerosing cholangitis. Endoscopy 1999;31:305–9. [DOI] [PubMed] [Google Scholar]
- 31.van Heek T, Rader AE, Offerhaus GJ, et al. K-ras, p53, and DPC4 (MAD4) alterations in fine-needle aspirates of the pancreas: a molecular panel correlates with and supplements cytologic diagnosis. Am J Clin Pathol 2002;117:755–65. [DOI] [PubMed] [Google Scholar]
- 32.Tada M, Komatsu Y, Kawabe T, et al. Quantitative analysis of K-ras gene mutation in pancreatic tissue obtained by endoscopic ultrasonography-guided fine needle aspiration: clinical utility for diagnosis of pancreatic tumor. Am J Gastroenterol 2002;97:2263–70. [DOI] [PubMed] [Google Scholar]
- 33.Kondo H, Sugano K, Fukayama N, et al. Detection of K-ras gene mutations at codon 12 in the pancreatic juice of patients with intraductal papillary mucinous tumors of the pancreas. Cancer 1997;79:900–5. [DOI] [PubMed] [Google Scholar]
- 34.Furuya N, Kawa S, Akamatsu T, et al. Long-term follow-up of patients with chronic pancreatitis and K-ras gene mutation detected in pancreatic juice. Gastroenterology 1997;113:593–8. [DOI] [PubMed] [Google Scholar]
- 35.Song MM, Nio Y, Sato Y, et al. Clinicopathological significance of Ki-ras point mutation and p21 expression in benign and malignant exocrine tumors of the human pancreas. Int J Pancreatol 1996;20:85–93. [DOI] [PubMed] [Google Scholar]
- 36.Hruban RH, Goggins M, Parsons J, et al. Progression model for pancreatic cancer. Clin Cancer Res 2000;6:2969–72. [PubMed] [Google Scholar]
- 37.Hruban RH, Wilentz RE, Goggins M, et al. Pathology of incipient pancreatic cancer. Ann Oncol 1999;10:9–11. [PubMed] [Google Scholar]
- 38.Klein WM, Hruban RH, Klein-Szanto AJ, et al. Direct correlation between proliferative activity and dysplasia in pancreatic intraepithelial neoplasia (PanIN): additional evidence for a recently proposed model of progression. Mod Pathol 2002;15:441–7. [DOI] [PubMed] [Google Scholar]
- 39.Berthelemy P, Bouisson M, Escourrou J, et al. Identification of K-ras mutations in pancreatic juice in the early diagnosis of pancreatic cancer. Ann Intern Med 1995;123:188–91. [DOI] [PubMed] [Google Scholar]
- 40.Brat DJ, Lillemoe KD, Yeo CJ, et al. Progression of pancreatic intraductal neoplasias to infiltrating adenocarcinoma of the pancreas. Am J Surg Pathol 1998;22:163–9. [DOI] [PubMed] [Google Scholar]
- 41.Tersmette AC, Petersen GM, Offerhaus GJ, et al. Increased risk of incident pancreatic cancer among first-degree relatives of patients with familial pancreatic cancer. Clin Cancer Res 2001;7:738–44. [PubMed] [Google Scholar]
- 42.Hruban RH, Petersen GM, Goggins M, et al. Familial pancreatic cancer. Ann Oncol 1999;10 (suppl 4) :69–73. [PubMed] [Google Scholar]
- 43.Lynch HT, Deters CA, Lynch JF, et al. Challenging pancreatic cancer-prone pedigrees: a nosologic dilemma. Am J Gastroenterol 2002;97:3062–70. [DOI] [PubMed] [Google Scholar]
- 44.Lynch HT, Brand RE, Lynch JF, et al. Hereditary factors in pancreatic cancer. J Hepatobiliary Pancreat Surg 2002;9:12–31. [DOI] [PubMed] [Google Scholar]
- 45.Brentnall TA. Cancer surveillance of patients from familial pancreatic cancer kindreds. Med Clin North Am 2000;84:707–18. [DOI] [PubMed] [Google Scholar]
- 46.Brentnall TA, Bronner MP, Byrd DR, et al. Early diagnosis and treatment of pancreatic dysplasia in patients with a family history of pancreatic cancer. Ann Intern Med 1999;131:247–55. [DOI] [PubMed] [Google Scholar]
- 47.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, et al. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol 2002;160:1239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]