Abstract
The damage-inducible UmuD′ and UmuC proteins are required for most SOS mutagenesis in Escherichia coli. Our recent assay to reconstitute this process in vitro, using a native UmuD′2C complex, revealed that the highly purified preparation contained DNA polymerase activity. Here we eliminate the possibility that this activity is caused by a contaminating DNA polymerase and show that it is intrinsic to UmuD′2C. E. coli dinB has recently been shown to have DNA polymerase activity (pol IV). We suggest that UmuD′2C, the fifth DNA polymerase discovered in E. coli, be designated as E. coli pol V. In the presence of RecA, β sliding clamp, γ clamp loading complex, and E. coli single-stranded binding protein (SSB), pol V’s polymerase activity is highly “error prone” at both damaged and undamaged DNA template sites, catalyzing efficient bypass of abasic lesions that would otherwise severely inhibit replication by pol III holoenzyme complex (HE). Pol V bypasses a site-directed abasic lesion with an efficiency about 100- to 150-fold higher than pol III HE. In accordance with the “A-rule,” dAMP is preferentially incorporated opposite the lesion. A pol V mutant, UmuD′2C104 (D101N), has no measurable lesion bypass activity. A kinetic analysis shows that addition of increasing amounts of pol III to a fixed level of pol V inhibits lesion bypass, demonstrating that both enzymes compete for free 3′-OH template-primer ends. We show, however, that despite competition for primer-3′-ends, pol V and pol III HE can nevertheless interact synergistically to stimulate synthesis downstream from a template lesion.
Although cellular DNA damage results in the induction of at least 30–40 proteins as part of the global SOS response in Escherichia coli (1–3), only two LexA-regulated genes, umuD and umuC, are required to observe DNA damage-induced mutagenesis (4). The Umu-dependent process of translesion DNA synthesis allows cells to survive in the presence of relatively high levels of replication-inhibiting DNA damage, but at the cost of an increased mutation rate (5, 6). Damage-inducible UmuD2 does not facilitate translesion DNA synthesis; under certain conditions, it inhibits it (7). Translesion DNA synthesis is achieved after UmuD2 undergoes a RecA-mediated intermolecular cleavage reaction (8) generating a shorter active UmuD′2 protein (9–11). The combination of UmuD′2 with UmuC forms a mutagenically active UmuD′2C complex that is stable and resistant to proteolytic degradation (12–14).
The current “working” model for SOS-induced mutagenesis posits that pol III holoenzyme complex (HE) becomes stalled at a DNA template lesion, at which point UmuD′2C binds the tip of an activated RecA protein (RecA*)-coated DNA filament, bringing the Umu mutagenic complex in contact with the stalled pol III complex (15). Genetic data suggested that pol III might incorporate a nucleotide opposite a lesion while Umu could then be involved in shepherding the polymerase past the damaged template site (16, 17).
Recently, we reconstituted an SOS mutagenesis assay in vitro using UmuD′2C purified as a native soluble complex (12) to copy a site directed abasic template lesion (18). Guided by the genetic data, we anticipated that lesion bypass would require the concerted action of at least three protein complexes—UmuD′2C, RecA*, and pol III HE. Indeed, we found that vigorous UmuD′2C-dependent lesion bypass occurred in the presence of pol III HE, also requiring RecA* (18).
However, we were surprised to find that the bypass reaction appeared equally efficient when pol III core was omitted from the reaction, prompting us to suggest that UmuD′2C might be an “error-prone” E. coli DNA polymerase (18). Thus, although we have proposed that “Umu might contain a distinct polymerizing component” (18), the unlikely possibility remained that the incorporation errors observed at normal and aberrant template sites might be caused by the presence of a minute pol III or pol II contaminant whose properties were altered in the presence of UmuD′2C. In this paper, we provide data that eliminate the possibility that UmuD′2C is contaminated by either pol III or pol II and confirm that UmuD′2C is a bona fide “error-prone” DNA polymerase (E. coli pol V). E. coli DinB protein has recently been shown to have DNA polymerase activity and has been designated as pol IV (19).
MATERIALS AND METHODS
Materials.
Ultrapure ATP and deoxyribonucleoside triphosphates were purchased from Amersham Pharmacia, and [γ-32P]ATP (4,000 Ci/mmol; 1 Ci = 37 GBq) was obtained from ICN. Purification of pol III and its accessory proteins was carried out as described (20). Pol I antibody was a generous gift from Lawrence Loeb (University of Washington, Seattle). E. coli single-stranded binding protein (SSB) and RecA protein were purchased from Amersham Pharmacia. Other materials have been described previously (18).
Strains and Plasmids.
Although UmuD′2C has been highly purified, we could not entirely rule out that its associated polymerase activity was caused by low levels of pol II or pol III (18). To address this issue, we have constructed a strain, RW588, carrying a deletion of the polB locus and a mutation (dnaE1026) rendering pol III temperature sensitive (21, 22). RW588 (ΔumuDC596∷ermGT ΔpolBΩspc dnaE1026 uvrA6) was constructed by P1 transduction of the dnaE1026 allele into DV08 (23), by selecting for Pro+ transductants and screening for dnaE1026-associated temperature sensitivity. The F′IQ from DH5αF′IQ (GIBCO/BRL) was transferred into RW588 and the strain transformed with the ptacUmuD′C overproducing plasmid, pOS1 (12). A second approach was to purify a mutant UmuD′2C by subcloning the umuC104 allele (6, 24) as an MluI-HindIII fragment from pRW124–104 (25) into a similarly digested pOS1 vector to generate pRW392.
Partial Purification of UmuD′2C Complex from a dnaE1026 ΔpolB Strain.
The earlier purification steps were carried out as described (18). In addition, the cell debris obtained after cell lysis was reextracted with R buffer (20 mM Tris⋅HCl, pH7.5/0.1 mM EDTA/1 mM DTT/20% glycerol) with 1 M NaCl and precipitated with polyethyleneimine (1.1% wt/vol). The lysate and supernatant containing UmuD′2C were combined and precipitated twice with ammonium sulfate (18). Proteins from the second ammonium sulfate precipitation were centrifuged, resuspended, and dialyzed in R buffer containing 1 M NaCl. The dialyzed proteins were loaded onto a Superdex 200 size exclusion column (16/60, Amersham Pharmacia) and run in R buffer with 1 M NaCl, but containing no glycerol. Fractions (1.1 ml) were collected, and glycerol was added to individual fractions to reach a final concentration of 20%. Fractions containing UmuD′2C proteins were visualized by staining with Coomassie blue R-250, resolved on a 12% SDS/PAGE gel. Fractions containing pol III temperature sensitive (ts) were detected by Western blotting by using antiserum directed against α subunit. The mutant UmuD′2C104 (D101N) protein, isolated from a wild-type background, was purified in a similar manner except that gel filtration was carried out by using Superdex 75 (26/60, Amersham Pharmacia), followed by phosphocellulose column chromatography (12).
Replicative Bypass Assay.
Translesion bypass reactions were carried out by using a 32P-labeled primer annealed to a linear 7.2-kb M13 DNA template with an abasic lesion, X, located 50 bases from the 5′-end, as described in ref. 18, with slight modifications. BSA (40 μg/ml) was added to the reactions, and polyethylene glycol was added to a final concentration of 5% (vol/vol) in a subset of reactions as indicated in the figure legends. Preincubation of UmuD′2C with other proteins was for 3 min. All reactions were performed in the presence of pol I antibody, which completely neutralized pol I activity. The dNTP concentrations were held constant at 100 μM each, unless specified otherwise.
Kinetic Analysis of Nucleotide Incorporation Efficiency Opposite an Abasic Lesion.
A gel kinetic analysis was used to determine the rate of abasic translesion synthesis as a function of dNTP concentration (26). Elongated 32P-labeled primer extension products were separated by using 16% polyacrylamide denaturing gels. Integrated polyacrylamide gel band intensities were measured with a PhosphorImager by using imagequant software (Molecular Dynamics). The efficiency of lesion bypass is determined by measuring IXΣ/IX−1, where IXΣ designates the integrated gel band intensities of primers extended opposite the abasic site, X, and beyond, i.e., translesion synthesis, and IX−1 is the integrated gel band intensity of primers extended by addition of a single running-start nucleotide (C) opposite the template site before X (26). Saturation plots of velocity as a function of dNTP concentration were carried out by using sigma plot software (Jandel, San Rafael, CA). Apparent Vmax and Km values were obtained from nonlinear least squares fits to a rectangular hyperbola, from a linear least squares fit to the “linear” region or from double reciprocal Lineweaver–Burk plots.
RESULTS
The experiments in this paper are focused primarily on determining whether UmuD′2C has an intrinsic DNA polymerase activity separate from pol II and pol III, even though it requires the same auxiliary proteins (β sliding clamp and γ clamp loading complex) for optimal activity (18). Our earlier study, showing that UmuD′2C copies past abasic template lesions in the absence of exogenous DNA polymerase (18), contained the caveat that UmuD′2C might contain trace contaminants of either pol III core, α subunit, or perhaps pol II. To eliminate this possibility, we have now purified UmuD′2C from an E. coli mutant strain containing a deletion of pol II (ΔpolB) (27) and a temperature-sensitive allele of DNA polymerase III (dnaE1026) (28).
UmuD′2C Is a DNA Polymerase, E. coli pol V.
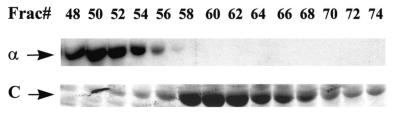
Cell lysates were enriched for soluble UmuD′2C complex by precipitation with polyethyleneimine and then with ammonium sulfate (12, 18). The UmuD′2C-enriched fraction was run on a high-resolution Superdex 200 column, and individual fractions were assayed for the presence of pol III α subunit by Western blots (Fig. 1 Upper) and for UmuC by using a Coomassie-stained gel (Fig. Lower). Western blots with polyclonal antibodies directed against UmuC and UmuD′ showed that the fractions containing the UmuD′2C complex correspond precisely to the locations determined with the Coomassie-stained gel (data not shown).
Figure 1.
Separation of pol IIIts from the UmuD′2C complex by using Superdex 200 gel filtration. (Upper) Superdex 200 fractions (30 μl) containing pol III α subunit [designated as α (Left)] were resolved on an 8% SDS/PAGE gel and visualized by chemiluminescent immunodetection by using antiserum directed against α subunit. (Lower) Superdex 200 fractions (20 μl) containing UmuC protein [designated as C (Left)] were visualized on a 12% SDS/PAGE gel stained with Coomassie blue R-250. The presence of UmuC and UmuD proteins was verified by using UmuC and UmuD′ antisera (data not shown).
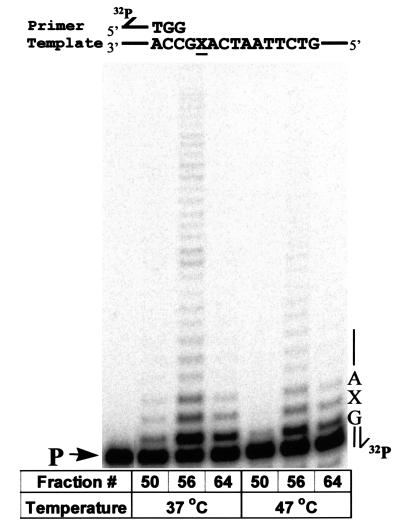
Three Superdex fractions were chosen to measure polymerase and abasic lesion bypass activities: fraction (fx) 50 contains predominantly pol IIIts core with a small amount of UmuD′2C; fx 56 contains overlapping pol IIIts + UmuD′2C; fx 64 contains UmuD′2C having no detectable pol IIIts present (Fig. 2). The lesion bypass assay (7, 18) is performed by using a 32P-labeled 30-mer primer annealed to a linear M13 DNA template containing a single abasic lesion located two positions downstream from the primer-3′-end (Fig. 2). Primer extension is carried out with each of the three fractions in the presence of RecA, β, γ complex, SSB, 4 dNTP substrates, and ATP. Extended primers are separated according to length by PAGE and visualized by PhosphorImaging. A single running-start nucleotide (dCMP) is incorporated opposite template G to reach the abasic lesion X. A primer band extended directly opposite X corresponds to nucleotide incorporation directly opposite the lesion. Longer primer extension bands reflect synthesis proceeding past the lesion site, i.e., lesion bypass.
Figure 2.
UmuD′2C (E. coli polV) has DNA polymerase activity. UmuD′2C was purified from a ΔpolB dnaE1026ts strain, RW588. Sephadex 200 fractions fx (50) containing predominantly pol IIIts; fx (56) containing pol IIIts + UmuD′2C and fx (64) containing UmuD′2C having no detectable pol IIIts were assayed for polymerase activity by extension of a 32P-labeled 30-mer primer annealed to a linear M13 DNA template, at permissive (37°C) and nonpermissive (47°C) temperatures. A single running-start base, C, is incorporated opposite G to reach the abasic lesion, X. The left-hand lane contains the primer (P) in the absence of proteins. Each reaction mixture contains 1.5 μl of the indicated fraction from a Superdex 200 gel filtration column (Fig. 1). All reactions were carried out in the presence of pol I antibody, RecA, SSB, β, γ complex, 4 dNTPs, and ATP.
At the permissive temperature (37°C), fx 50 (containing pol IIIts) incorporates a running-start C opposite G along with a small amount of incorporation opposite X and 1 nt beyond, but all synthesis is essentially absent at the nonpermissive temperature of 47°C (Fig. 2). The extremely weak lesion bypass band observed at 37°C can be attributed to the presence of a small amount of UmuD′2C (Fig. 1; see e.g. ref. 18). In contrast, fx 64, which contains no detectable pol IIIts, carries out running-start incorporation and lesion bypass at both permissive and nonpermissive temperatures (Fig. 2). We conclude, therefore, that UmuD′2C has its own polymerase activity that can incorporate a running-start C opposite G in a template-directed reaction and can also catalyze nucleotide incorporation and continued extension at an abasic template lesion.
Polymerization carried out by fx 56 (UmuD′2C + pol IIIts) revealed an interesting result. The robust incorporation and lesion bypass observed at 37°C is consistent with our previous observation that UmuD′2C catalyzes incorporation directly opposite X and then extends several bases beyond the lesion, at which point UmuD′2C dissociates and is replaced by pol III HE (18). However, unexpectedly, this synergistic effect on downstream synthesis also occurs at the nonpermissive temperature. As seen in Fig. 2, the amount of total DNA synthesis is much greater for fx 56 (UmuD′2C + pol IIIts) compared with essentially no DNA synthesis for fx 50 (pol IIIts) and moderate synthesis for fx 64 (UmuD′2C). It appears that the temperature-sensitive mutant pol III is stabilized in the presence of high concentrations of UmuD′2C, suggesting that the two polymerases might interact in the vicinity of the lesion. Perhaps once lesion bypass has occurred, normal DNA replication can resume by replacing a distributive Umu polymerase (18) with a processive pol III HE.
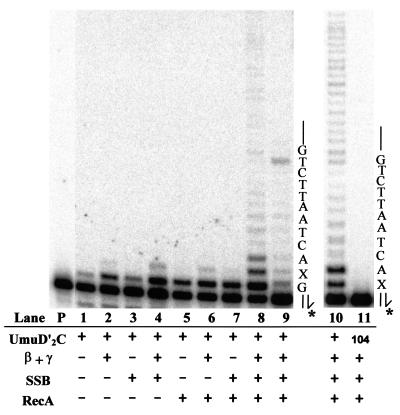
We investigated requirements for the other protein cofactors by permuting RecA, β, γ complex, and SSB in the lesion bypass reactions (Fig. 3). We observed that UmuD′2C alone clearly catalyzes incorporation of the running-start C but cannot measurably catalyze further extension opposite the lesion (lane 1). Inclusion of either β, γ complex (lane 2), SSB (lane 3), or RecA (lane 5) stimulates incorporation of C, but there is essentially no incorporation opposite X. Weak UmuD′2C-catalyzed incorporation opposite X occurs in the presence of β, γ complex + either SSB (lane 4) or RecA (lane 6), but lesion bypass is not observed. However, a strong lesion bypass signal was observed once RecA was included along with these other three components (lane 8). Indeed, RecA appears to be the most important stimulatory factor required for translesion synthesis by UmuD′2C.
Figure 3.
Protein cofactor requirements for UmuD′2C (E. coli pol V)-catalyzed lesion bypass. Each reaction in lanes 1–9 contains 1.5 μl of Superdex 200 fx 64 containing UmuD′2C (having no detectable pol IIIts), pol I antibody, and various combinations of RecA, β, γ complex, and SSB, as indicated (lanes 1–9). Running-start reactions, in which C is incorporated opposite template G to reach the abasic site, were carried out at 37°C, with all four dNTPs present in lanes 1 to 8, but with dCTP omitted in lane 9. Reactions were run in the presence of 5% polyethylene glycol. A portion of the template sequence is shown at the right of lane 9, where X represents an abasic site. The left-hand lane contains the primer (P) in the absence of proteins. Standing-start reactions, in which the first incorporated nucleotide occurs opposite X, were run for wild-type UmuD′2C (lane 10) and for the mutant UmuD′2C104 (D101N) (lane 11), each at a concentration of 200 nM. A portion of the template, shown at the right of lane 11, has the same sequence as the running-start template, but uses a primer terminating one base before the lesion. The asterisk (*) designates a 32P-label at the 5′-primer terminus. The running-start primer-template DNA is shown at the top of Fig. 2.
Primer extension was reduced significantly when dCTP was omitted from the reaction (lane 9). The small amount of synthesis occurring in the absence of dCTP can be attributed to UmuD′2C-catalyzed misincorporation found to occur at undamaged template sites (18). Note the presence of a termination band appearing before the second template G (lane 9) that disappears when all four dNTPs are present (lane 8). These data demonstrate that UmuD′2C is behaving as a DNA polymerase, not a terminal deoxynucleotidyl transferase. A mutant UmuD′2C complex containing UmuC104 (D101N) cannot copy past the abasic lesion (lane 11), thereby confirming that the error-prone polymerase activity is intrinsic to UmuD′2C and cannot be explained by contamination with some other polymerase, e.g., the recently discovered pol IV (DinB) (19).
UmuD′2C Preferentially Incorporates A Opposite an Abasic Lesion and Competes with pol III for Binding Primer-3′OH-Ends.
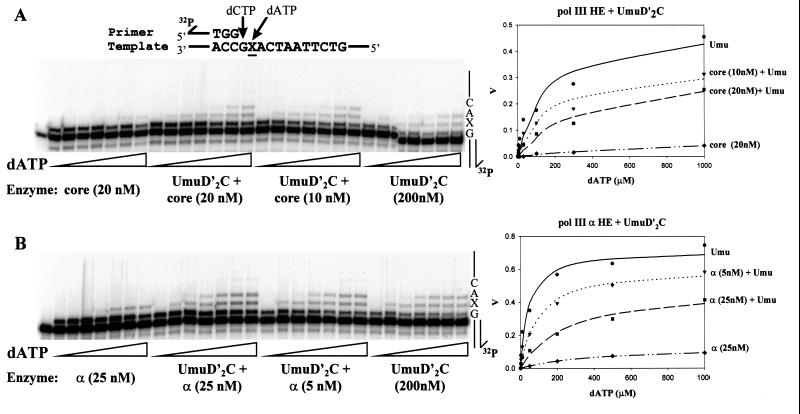
Kinetic experiments were performed to determine how the presence of pol III influences UmuD′2C-catalyzed lesion bypass (Fig. 4). A running-start reaction, in which a single nucleotide (C) is incorporated before reaching the abasic lesion, was used to measure incorporation directly opposite and downstream from the abasic lesion as a function of dATP substrate concentration. Two pol III HE forms were used, one containing the proofreading-proficient pol III core (α, ɛ, and θ subunits) (Fig. 4A) and the second form containing α, in the absence of ɛ and θ (Fig. 4B). RecA, β sliding clamp, γ clamp loading complex, and SSB were present in all reactions.
Figure 4.
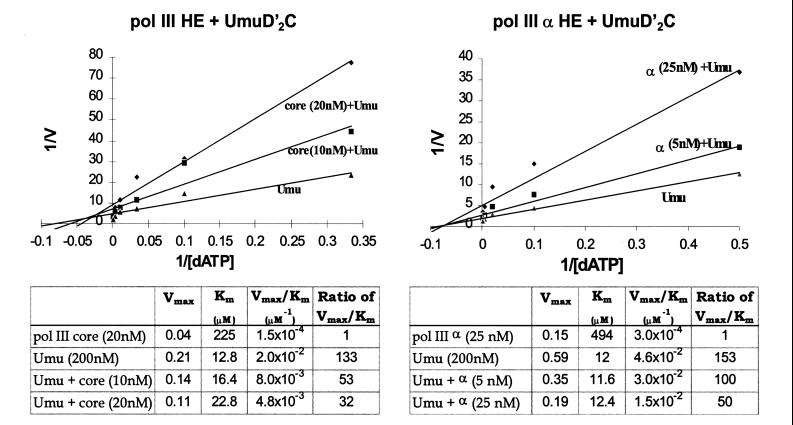
Inhibition of UmuD′2C (E. coli pol V)-catalyzed translesion synthesis by wild-type pol III. A running-start C (20 μM dCTP) is incorporated opposite template G before reaching the abasic lesion, X. The concentration of dATP used for incorporation opposite X was varied to measure the kinetics of incorporation by either UmuD′2C, wild-type pol III, or a combination of both proteins. The UmuD′2C complex used in the reactions was purified from a strain containing wild-type pol III (18). Michaelis–Menten saturation plots of the translesion synthesis rate vs. dATP concentration are shown at the right. The translesion synthesis rate V is obtained by measuring IXΣ/IX−1 as a function of dATP concentration, where IXΣ are the integrated gel band intensities for incorporation at the site of the lesion and beyond, and IX−1 is the integrated gel band intensity at the G site, before reaching the lesion (see Materials and Methods). (A) Translesion synthesis catalyzed by UmuD′2C, pol III core HE, or combinations of both. (B) Translesion synthesis catalyzed by UmuD′2C, pol III α HE, or combinations of both. The dATP concentrations used were 0, 3, 10, 30, 100, 300, and 1,000 μM for A and 0, 2, 10, 50, 200, 500, and 1,000 μM for B. UmuD′2C is present at 200 nM in each experiment. The reactions were run at 37°C in the presence of RecA, SSB, and β, γ complex.
In the absence of pol III, UmuD′2C incorporates A opposite X, and further extension to the next template site occurs by incorporation of an A⋅A mispair (Fig. 4). In contrast, α HE incorporates a much lower amount of A opposite X with no observable bypass, while pol III HE shows hardly any incorporation of A even at high dATP concentration (300 μM).
A key result is that the rate of UmuD′2C-catalyzed lesion bypass is reduced significantly as the concentration of either of the two pol III HE forms is increased. It is important to emphasize that we are measuring lesion bypass synthesis as the amount of incorporation opposite the lesion and extension past the lesion, normalized to the amount of primer extended by incorporation of running-start C to reach the lesion site. Although incorporation of running-start C just before the lesion is strongly stimulated as the concentration of pol III is increased relative to UmuD′2C, the efficiency of translesion synthesis is reduced substantially. In other words, the presence of pol III HE at the primer-3′-end prevents UmuD′2C from binding at the lesion site.
A double reciprocal plot of the data is used to illustrate this important point (Fig. 5). We observe that the apparent Km values for incorporation of A opposite X and opposite the downstream A are relatively insensitive to the concentration of either pol III or α holoenzymes. The apparent Km values are roughly the same compared with UmuD′2C alone (≈ 12 to 23 μM) and are approximately 20- and 40-fold lower than for the pol III HE and α HE, respectively (Fig. 5). This apparent noncompetitive inhibition of UmuD′2C by pol III, with respect to dNTP incorporation at X, implies that UmuD′2C and pol III are competing for the same primer-3′-ends. Thus, whenever UmuD′2C succeeds in binding to a primer-end and catalyzes bypass synthesis in the presence of pol III, it does so with a characteristically low Km value. Conversely, large reductions in Vmax occur because there are fewer bound UmuD′2C-primer-template DNA complexes because of competition by pol III.
Figure 5.
Kinetic analysis of the effect of wild-type pol III on UmuD′2C (E. coli pol V)-catalyzed translesion synthesis. Lineweaver–Burk double reciprocal plots for pol III HE and α HE (Upper) were generated from the kinetic data given in Fig. 4. Apparent Km and Vmax values and their ratios characterizing translesion synthesis by UmuD′2C (designated as Umu) in the absence and presence of pol III are provided in the tables below.
The kinetic data reveal two other important points: (i) UmuD′2C catalyzes bypass synthesis with at least 100-fold higher efficiency than pol III, either with or without proofreading; (ii) A is favored for incorporation opposite the abasic lesion by roughly 3-fold over G (data not shown), in accordance with the “A-rule” (29). Incorporation of either C or T opposite X is barely detectable (data not shown).
DISCUSSION
When pol III HE was used in an earlier study to copy a site-directed abasic template lesion in vitro, the two aberrant polymerase reactions, insertion of a nucleotide opposite the lesion and bypass of the lesion by incorporating a “next-correct” nucleotide, were barely detectable (18). But both reactions were stimulated strongly when UmuD′2C, RecA, and SSB were included in the reaction. Remarkably, however, omitting pol III core had essentially no effect on lesion bypass. These data led us to suggest that UmuD′2C is an error-prone DNA polymerase that incorporates a nucleotide opposite a damaged template site and then synthesizes past the lesion (18). However, we could not definitively rule out that lesion bypass might be caused alternatively by contamination with minute amounts of pol III or pol II exhibiting drastically reduced fidelity in the presence of UmuD′2C.
UmuD′2C Is an Error-Prone DNA Polymerase, E. coli pol V.
Several lines of evidence are presented in this paper demonstrating that UmuD′2C has a polymerase activity distinct from pol III or pol II. We purified an intact native UmuD′2C complex from cells lacking pol II and containing a temperature-sensitive mutant pol III. UmuD′2C and pol IIIts were separated by Superdex 200 gel filtration (Fig. 1), and three column fractions were used to investigate bypass of a site-directed abasic lesion (Fig. 2). One fraction (fx 50) contained the pol IIIts peak along with a small amount of UmuD′2C, while another (fx 64) contained UmuD′2C peak with no detectable pol IIIts (Fig. 1). We also investigated the properties of a third fraction (fx 56) enriched for UmuD′2C but having a small, yet significant, amount of pol IIIts.
In the assay, 32P-labeled primer is extended by adding a single running-start nucleotide (dCTP) before reaching an abasic template site, X. In contrast to pol IIIts (fx 50), which incorporates C opposite G at 37°C, but whose activity is reduced substantially at 47°C, UmuD′2C (fx 64) incorporates C and catalyzes lesion bypass at both permissive and nonpermissive temperatures (Fig. 2). Thus, UmuD′2C′s polymerase activity cannot be explained by the presence of pol III. Contamination with pol II is ruled out because a ΔpolB strain was used to overexpress UmuD′2C, and pol I is ruled out because the assays were performed in the presence of large excess of neutralizing antibody directed against pol I. We also verified that UmuD′2C104 fails to catalyze lesion bypass (Fig. 3, lane 11), demonstrating that the data cannot be attributed to any other polymerase such as pol IV (DinB) (19).
Accessory Proteins Required for UmuD′2C-catalyzed Lesion Bypass.
UmuD′2C-catalyzed lesion bypass requires the presence of activated RecA (Fig. 3, ref. 18), in agreement with genetic data (11, 30, 31). However, we find that Umu-catalyzed bypass also requires the presence of β, γ complex and SSB, in addition to RecA (Fig. 3, lanes 9 and 10; ref. 18). Indeed, incorporation of a running-start nucleotide at an undamaged template site is stimulated in the presence of each of these accessory proteins (Fig. 3). Because UmuD′2C polymerase activity appears to be highly distributive, it will be interesting to determine whether the β processivity clamp might be responsible for stabilizing the Umu–DNA complex in the vicinity of a lesion.
Pol III and UmuD′2C Compete for Primer-3′-OH Ends.
Kinetic data show that UmuD′2C-catalyzed lesion bypass activity is reduced as the concentration of pol III is increased, a result that cannot be attributed to pol III proofreading (Fig. 4). The fact that catalysis by pol III at a lesion site is negligible compared with UmuD′2C enables a straightforward interpretation of the kinetic data. The observation that Vmax is reduced by addition of pol III while the apparent Km for lesion bypass is essentially the same as for UmuD′2C alone indicates that both enzymes are able to bind primer-3′-ends, providing strong independent evidence that UmuD′2C is a low-fidelity DNA polymerase.
How can these data be reconciled with genetics experiments that have shown a clear requirement for pol III in SOS mutagenesis (16, 32)? Our experiments show that UmuD′2C is not processive and is unlikely to be able to duplicate the 4.6-Mb E. coli chromosome, a task normally performed by pol III HE. Pol III HE must, therefore, take over from UmuD′2C once bypass has occurred. Indeed, our data showing that UmuD′2C appears to stabilize pol IIIts (Fig. 2) imply that pol III and UmuD′2C may physically interact near the site of a lesion. If this interpretation is valid, then one can visualize how UmuD′2C might displace pol III core blocked at template lesion, interact with β, RecA*, and SSB to copy past the lesion, and subsequently be released from the DNA to allow reentry of pol III several bases downstream.
UmuC belongs to a large superfamily of proteins that can be broadly subclassified into UmuC-like, DinB-like, Rev1-like, and Rad30-like proteins (33, 34). Our finding that UmuD′2C is a DNA polymerase means that the prototype for each subfamily has been shown to possess the ability to catalyze phosphodiester bond formation. UmuC, DinB, and Rad30 appear to be bona fide polymerases (this paper and refs. 19, 35), whereas Rev1 is a template-directed dCTP transferase (36). One unusual characteristic of UmuD′2C, however, is its complex dependence on a diverse group of other proteins, RecA*, β, γ complex, and SSB to perform its main function in copying past replication blocking lesions. The availability of a reconstituted lesion bypass assay in vitro (18, 37) should permit the elucidation of each of these interactions and their specific roles in translesion DNA synthesis.
Acknowledgments
We thank Dr. Phuong Pham for help in purifying UmuD′2C104. This work was supported by National Institutes of Health Grants GM42554 and GM38839.
ABBREVIATIONS
- RecA*
activated RecA protein
- pol
polymerase
- HE
holoenzyme complex
- SSB
E. coli single-stranded binding protein
- fx
Superdex 200 column fraction
- ts
temperature sensitive
References
- 1.Witkin E M. Bacteriol Rev. 1976;40:869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 1995. [Google Scholar]
- 3.Koch W H, Woodgate R. In: The SOS Response. Nickoloff J A, Hoekstra M F, editors. Totowa, NJ: Humana; 1988. pp. 107–134. [Google Scholar]
- 4.Sommer S, Knezevic J, Bailone A, Devoret R. Mol Gen Genet. 1993;239:137–144. doi: 10.1007/BF00281612. [DOI] [PubMed] [Google Scholar]
- 5.Kato T, Shinoura Y. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- 6.Steinborn G. Mol Gen Genet. 1978;165:87–93. doi: 10.1007/BF00270380. [DOI] [PubMed] [Google Scholar]
- 7.Rajagopalan M, Lu C, Woodgate R, O’Donnell M, Goodman M F, Echols H. Proc Natl Acad Sci USA. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald J P, Peat T S, Levine A S, Woodgate R. J Mol Biol. 1999;285:2199–2209. doi: 10.1006/jmbi.1998.2433. [DOI] [PubMed] [Google Scholar]
- 9.Shinagawa H, Iwasaki T, Kato T, Nakata A. Proc Natl Acad Sci USA. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burckhardt S E, Woodgate R, Scheuremann R H, Echols H. Proc Natl Acad Sci USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nohmi T, Battista J R, Dodson L A, Walker G C. Proc Natl Acad Sci USA. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruck I, Woodgate R, McEntee K, Goodman M F. J Biol Chem. 1996;271:10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- 13.Frank E G, Gonzalez M, Ennis D G, Levine A S, Woodgate R. J Bacteriol. 1996;178:3550–3556. doi: 10.1128/jb.178.12.3550-3556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank E G, Gonzalez M, Ennis D G, Levine A S, Woodgate R. Proc Natl Acad Sci USA. 1996;93:10291–10296. doi: 10.1073/pnas.93.19.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sommer S, Boudsocq F, Devoret R, Bailone A. Mol Microbiol. 1998;28:281–291. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 16.Hagensee M E, Timme T L, Bryan S K, Moses R E. Proc Natl Acad Sci USA. 1987;84:4195–4199. doi: 10.1073/pnas.84.12.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bridges B A, Woodgate R. Proc Natl Acad Sci USA. 1985;82:4193–4197. doi: 10.1073/pnas.82.12.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang M, Bruck I, Eritja R, Turner J, Frank E G, Woodgate R, O’Donnell M, Goodman M F. Proc Natl Acad Sci USA. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner, J., Gruz, P., Su-Ryang, K., Uamada, M., Matsui, K., Fuchs, R. P. P. & Nohmi, T. (1999) Mol. Cell., in press. [DOI] [PubMed]
- 20.Naktinis V, Turner J, O’Donnell M. Cell. 1996;84:137–145. doi: 10.1016/s0092-8674(00)81000-4. [DOI] [PubMed] [Google Scholar]
- 21.Wechsler J A, Gross J D. Mol Gen Genet. 1971;113:273–284. doi: 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
- 22.Gefter M L, Hirota Y, Kornberg T, Wechsler J A, Barnoux C. Proc Natl Acad Sci USA. 1971;68:3150–3153. doi: 10.1073/pnas.68.12.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandewiele D, Borden A, O’Grady P I, Woodgate R, Lawrence C. Proc Natl Acad Sci USA. 1998;95:15519–15524. doi: 10.1073/pnas.95.26.15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch W H, Ennis D G, Levine A S, Woodgate R. Mol Gen Genet. 1992;233:443–448. doi: 10.1007/BF00265442. [DOI] [PubMed] [Google Scholar]
- 25.Woodgate R, Singh M, Kulaeva O I, Frank E G, Levine A S, Koch W H. J Bacteriol. 1994;176:5011–5021. doi: 10.1128/jb.176.16.5011-5021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creighton S, Bloom L B, Goodman M F. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 27.Escarcellar M, Hicks J, Gudmundsson G, Trump G, Touati D, Lovett S, Foster P, McEntee K, Goodman M F. J Bacteriol. 1994;176:6221–6228. doi: 10.1128/jb.176.20.6221-6228.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wechsler J A, Nusslein V, Otto B, Klein A, Bonhoeffer F, Herrmann R, Gloger L, Schaller H. J Bacteriol. 1973;113:1381–1388. doi: 10.1128/jb.113.3.1381-1388.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strauss B S. BioEssays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 30.Dutreix M, Moreau P L, Bailone A, Galibert F, Battista J R, Walker G C, Devoret R. J Bacteriol. 1989;171:2415–2413. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sweasy J B, Witkin E M, Sinha N, Roegner-Maniscalco V. J Bacteriol. 1990;172:3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bridges B A. Mutat Res. 1988;198:343–350. doi: 10.1016/0027-5107(88)90012-7. [DOI] [PubMed] [Google Scholar]
- 33.McDonald J P, Levine A S, Woodgate R W. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roush A A, Suarez M, Friedberg E C, Radman M, Seide W. Mol Gen Genet. 1998;257:686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- 35.Johnson R E, Prakash S, Praskah L. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 36.Nelson J R, Lawrence C W, Hinkle D C. Nature (London) 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 37.Reuven N B, Tomer G, Livneh Z. Mol Cell. 1998;2:191–199. doi: 10.1016/s1097-2765(00)80129-x. [DOI] [PubMed] [Google Scholar]