Abstract
The molecular mechanisms that determine the correct subcellular localization of proteins targeted to membranes by tail-anchor sequences are poorly defined. Previously, we showed that two isoforms of the tung oil tree [Vernicia (Aleurites) fordii] tail-anchored Cb5 (cytochrome b5) target specifically to ER (endoplasmic reticulum) membranes both in vivo and in vitro [Hwang, Pelitire, Henderson, Andrews, Dyer and Mullen (2004) Plant Cell 16, 3002–3019]. In the present study, we examine the targeting of various tung Cb5 fusion proteins and truncation mutants to purified intracellular membranes in vitro in order to assess the importance of the charged CTS (C-terminal sequence) in targeting to specific membranes. Removal of the CTS from tung Cb5 proteins resulted in efficient binding to both ER and mitochondria. Results from organelle competition, liposome-binding and membrane proteolysis experiments demonstrated that removal of the CTS results in spontaneous insertion of tung Cb5 proteins into lipid bilayers. Our results indicate that the CTSs from plant Cb5 proteins provide ER specificity by preventing spontaneous insertion into incorrect subcellular membranes.
Keywords: C-terminal sequence, cytochrome b5, endoplasmic reticulum, organelle specificity, tail-anchor protein, vesicle-associated membrane protein (VAMP)
Abbreviations: Cb5, cytochrome b5; RCb5, rat Cb5; CTS, C-terminal sequence; ER, endoplasmic reticulum; gPA, IgG-binding domain of Staphylococcus aureus protein A; NTS, N-terminal sequence; pOCT, preornithine carbamyl transferase; RCb5, rat Cb5; TMS, transmembrane sequence; Tom20, 20 kDa receptor subunit of the translocation complex of the outer mitochondrial membrane; VAMP, vesicle-associated membrane protein
INTRODUCTION
Proteins that contain a C-terminal membrane-binding sequence with the capacity for post-translational integration into membranes (a tail-anchor or insertion sequence) are involved in a wide variety of cellular functions including apoptosis, protein translocation, signal transduction and vesicle trafficking [1,2]. As a group, tail-anchored proteins have many different enzymatic properties including kinase, phosphatase and lipid biosynthesis activities and also contribute to mechanical functions such as permeabilization and fusion of membranes [3,4]. The membrane topology of tail-anchored proteins tethers a large functional cytoplasmic domain to the membrane; this has been equated to ordering a specific layer of cytoplasm close to the surface of organelles [2]. However, recent evidence suggests that some tail-anchored proteins may contain additional TMSs (transmembrane sequences) [5,6]. Most tail-anchored proteins characterized to date appear to target and insert into either of two distinct membrane systems: the mitochondrial outer membrane or the ER (endoplasmic reticulum). From the latter compartment they are sorted to other membranes such as the nuclear envelope, Golgi, vacuole/lysosome, peroxisomes, synaptic vesicles and plasma membrane [7,8].
The N-terminal targeting signals normally associated with mitochondrial and ER membrane proteins have not been found on tail-anchored proteins [9,10]. Instead, the signals for organelle-specific targeting appear to reside within the tail anchor sequence. For instance, the tail anchors of monoamine oxidase B, Bcl-2, VAMP1A (vesicle-associated membrane protein 1A), VAMP1B, and several different isoforms of rat and tung tree Cb5 (cytochrome b5), have been shown to contain all the necessary information for organelle-specific targeting of inert passenger proteins to ER, mitochondria or both organelles [2,11,12].
A typical tail anchor is composed of three regions: (i) the cytoplasmic NTS (N-terminal sequence), a central sequence of 15–22 hydrophobic amino acids (the TMS), and (ii) the CTS (C-terminal sequence). The NTS has been shown to contain targeting signals in some tail-anchored proteins [13,14]. Although referred to as the TMS, there is no inherent requirement that the central hydrophobic segment always traverses both leaflets of the bilayer as the functional definition of a tail anchor requires only that it anchors a protein to specific membranes [15]. The CTS includes all of the amino acid residues C-terminal of the TMS, is usually found within the target organelle lumen or matrix, and often contains targeting information [16–19].
Tail anchors containing positive charges that flank an amphipathic TMS are often recognized and inserted into the outer membrane of mitochondria [2,8,20]. The similarity of mitochondrial tail-anchor sequences to more traditional N-terminal mitochondrial signal anchors suggests that both signals may interact with the same shallow binding groove on the surface of Tom20 (the 20 kDa receptor subunit of the translocation complex of the outer mitochondrial membrane) [8,21–23]. Whether mitochondrial tail-anchor sequences interact with Tom20 or not, they actively direct proteins to mitochondria since their removal generally results in mislocalization of the truncated protein in the cytoplasm [11]. In some cases, removal of a mitochondria-specific CTS has been reported to result in targeting to the ER [9,17]. These latter observations are consistent with the ER being a default destination for some tail-anchor proteins. Signals for the targeting of tail-anchored proteins to ER were identified also in the NTS of VAMP1, VAMP2 and VAMP8 [14]. Mutation of positively charged amino acids or a conserved asparagine residue in these signals resulted in mislocalization of the mutant VAMP proteins to the cytosol. Therefore not all tail-anchored proteins default to the ER in response to disruption of their targeting signals.
Identification of targeting signals within the tail-anchor sequence of Cb5 has been difficult because the most commonly examined forms of the protein, the ER-specific isoforms of RCb5 (rat Cb5) or rabbit Cb5, target specifically to ER only in vivo. In cell-free assays, RCb5 targets to any membrane, including synthetic liposomes added to the reaction, and shows no preference for ER even in organelle competition experiments [12,24]. Previous examination of a mutant in which the charged CTS of rabbit Cb5 was replaced with a single threonine residue demonstrated insertion of the mutant into multiple membranes [16]. However, the change in targeting mechanism responsible could not be elucidated because rabbit Cb5 does not maintain targeting specificity in vitro. In contrast, recently identified tung tree Cb5 proteins, Cb5A and Cb5B, target exclusively to ER both in vivo in tobacco suspension-cultured cells and in vitro with purified ER or mitochondria derived from mammalian cells [11]. Because these Cb5 isoforms retain ER-specific targeting in vitro, we have used them here to identify a novel regulatory mechanism responsible for specificity in targeting to the ER. Our results show that removal of the CTS from the plant Cb5 isoforms did not inhibit targeting of these proteins to ER, but results in increased targeting efficiency at the expense of selectivity as the mutant protein also accumulated at mitochondria. This result indicates that the CTS is necessary for ER-specific targeting. Results from liposome-binding assays and experiments with proteolysed membranes suggest that the CTS of Cb5A conferred targeting specificity for ER by preventing spontaneous insertion of the TMS into bilayers. We propose that the CTSs from Cb5A and Cb5B prevent spontaneous insertion of the tail-anchor sequences of these proteins, thereby necessitating the involvement of proteinaceous machinery to bind them to membranes. Since these tail-anchor sequences are recognized at the ER but not recognized by the mitochondrial insertion machinery, the result is selective insertion into the ER. Thus our results demonstrate that organelle targeting specificity of a tail-anchored protein can be mediated by both specific recognition signals and by sequences that prevent spontaneous insertion into incorrect subcellular organelles.
EXPERIMENTAL
Plasmid construction
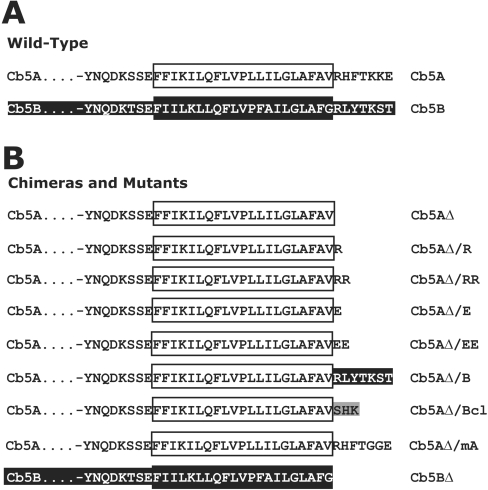
Construction of plasmids expressing either Cb5A, Myc–Cb5A, Cb5B, Bcl-2 or pOCTgPA (defined below) have been previously described [11,12,25]. The name assigned to each derivative of Cb5 reflects the sequence of the construct (Figure 1). The plasmid pSPUTK/Myc-Cb5B was constructed by ligating the NcoI–XbaI fragment from pRTL2/Myc-Cb5B [11] into NcoI–XbaI-digested pSPUTK. pSPUTK and pSPUTK-BglII contain the SP6 promoter and non-translated sequences that confer efficient translation to in vitro transcription products and have been described elsewhere [26,27]. The plasmids pSPUTK/Cb5AΔ and pSPUTK-BglII/Cb5BΔ were constructed using QuikChange® site-directed mutagenesis according to the manufacturer's instructions (Stratagene, La Jolla, CA, U.S.A.) with either pSPUTK/Cb5A or pSPUTK-BglII/Cb5B as template DNA and appropriate forward and reverse primers to replace the first codon of the Cb5A or Cb5B CTS (corresponding amino acid underlined, -RHFTKKE or -RLYTKST) with a stop codon. To modify sequences coding for residues in the CTS of Cb5A, site-directed mutagenesis was performed using pSPUTK/Cb5A as template DNA and appropriate primers to generate pSPUTK/Cb5AΔ/R, pSPUTK/Cb5AΔ/RR, pSPUTK/Cb5AΔ/E, pSPUTK/Cb5AΔ/EE, pSPUTK/Cb5AΔ/B, pSPUTK/Cb5AΔ/Bcl, or pSPUTK/Cb5AΔ/mA. Complete descriptions of all the oligonucleotide primers used in the construction of the plasmids used in the present study are available upon request. All constructs were verified by dideoxynucleotide sequencing.
Figure 1. Schematic representation of Cb5 constructs examined in the present study.
(A) Deduced amino acid sequences of the C-terminal portions of either wild-type Cb5A (black on white) or Cb5B (white on black). Amino acid sequences corresponding to the TMS in each of these three proteins are highlighted with oversized boxes. (B) Amino acid sequences in the C-termini of mutant Cb5A constructs indicated as in (A). Amino acids in the C-terminus of grey boxed residues were derived from Bcl-2 (Bcl).
Preparation of membranes
ER-enriched membrane vesicles, termed microsomes, were prepared from porcine pancreas as described by Walter and Blobel [28]. The resuspended microsomal pellet was then column washed [28]. Each batch of microsomes was tested for co-translational translocation of preprolactin [29]. One equivalent (Eq) of microsomes is approx. 1 μl of 50 A280 units/ml of rough ER membranes before salt extraction [28]. Mitochondria were isolated from rat liver as described by Greenawalt [30]. Isolated mitochondria and microsomes were assayed for purity by immunobloting for Sec61α, calreticulum, Hsp60 (heat-shock protein 60) and cytochrome c (Supplementary Figure 1, http://www.BiochemJ.org/bj/401/bj4010701add.htm). Phospholipid vesicles of mitochondrial-like lipid composition (molar percentage 48:28:10:10:4 phosphatidylcholine/phosphotidylethanolamine/phosphatidylserine/phosphatidylinositol/cardiolipin) were prepared by extrusion in 10 mM Tris/HCl (pH 7.5) [31]. Trypsin-treated microsomes were digested with 20 μg/ml of trypsin (Sigma–Aldrich, St Louis, MO, U.S.A.). After a 30 min incubation at 4 °C, PMSF was added to a final concentration of 2 mM to inactivate the protease. Trypsinized microsomes were then diluted and washed twice as previously described [32]. Mock-treated microsomes were processed the same way as the trypsin-treated microsomes but without addition of trypsin. Trypsin-treated mitochondria were prepared as previously described [12].
In vitro transcription, translation and membrane binding
In vitro transcription and translation were performed as described previously [29]. After incubation at 24 °C for 60 min, ribosomes were removed from the translation reactions by centrifugation at 200000 g for 10 min. An excess of membranes was added to the supernatants to ensure that targeting occurs in a post-translational manner and that membrane concentration is not limiting. Membrane binding experiments were conducted using 10 μl of the transcription–translation reaction incubated at 24 °C for 60 min, with either 1 μl of 1 Eq/1 μl microsomes or 25 μl of 2 μg/μl mitochondrial protein. In organelle competition experiments, three proteins were assayed simultaneously; therefore 30 μl of transcription–translation reaction was incubated with the indicated quantity of membranes at 24 °C for 60 min. Membrane binding assays [25] and organelle competition experiments were carried out as described by Hwang et al. [11]. Volumes of each fraction corresponding to equivalent amounts of the original translation reaction were analysed by SDS/PAGE using a Tris-Tricine buffer system [33]. Radioactive proteins were visualized and quantified using a phosphoimager and ImageQuant software (Amersham Biosciences, Piscataway, NJ, U.S.A.).
RESULTS
The most intensely studied Cb5 is the ER isoform from rat liver (RCb5). Unlike other tail-anchor proteins, in vitro RCb5 targets to virtually any membrane [12,24,34]. In contrast, two recently described isoforms of plant Cb5 (tung tree Cb5A and Cb5B) target to ER specifically both in vitro and in vivo ([11]; also Figure 2). To investigate the sequences within these proteins responsible for targeting fidelity in vitro, a series of mutants were examined in which sequences from the tail-anchored proteins Cb5A, Cb5B, and the anti-apoptosis protein Bcl-2 were interchanged (Figure 1).
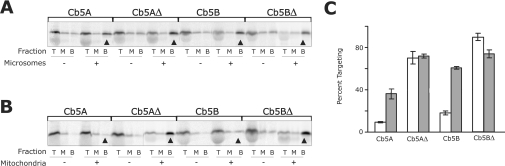
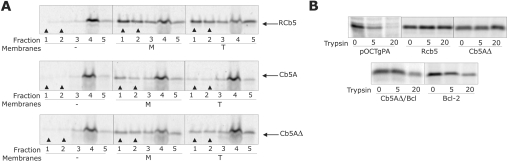
Figure 2. The CTSs of Cb5A and Cb5B are required for ER-specific targeting.
(A) Targeting of full-length and CTS-truncated versions of Cb5 isoforms A and B to ER membranes. Plasmids encoding Cb5A, Cb5AΔ, Cb5B or Cb5BΔ were transcribed in vitro using SP6 polymerase, and transcription products were translated in reticulocyte lysate containing [35S]methionine. After incubation at 24 °C for 60 min, ribosomes were removed from the translation reactions by centrifugation at 200000 g for 10 min. Translation products were then incubated with (+) or without (–) added microsomes. The targeting reactions were layered on top of a sucrose cushion, and the microsomes were pelleted by centrifugation for 11 min at 100000 g. The gradients were fractionated into top (T) and middle (M) fractions containing soluble proteins and the bottom (B) fraction containing microsomes and microsome-bound proteins (arrowheads). Equivalent amounts of all fractions were analysed by SDS/PAGE. (B) Targeting of full-length and CTS-truncated versions of Cb5 isoforms A and B to mitochondrial membranes. Plasmids were transcribed and translated as described in (A). Translation products were then incubated with (+) or without (–) added mitochondria, fractionated and analysed as in (A). (C) The percentage of the total protein recovered from the pellet fractions in (A) and (B) as determined using a phosphoimager. The percentage targeting reported is corrected for background by subtracting the percentage of protein in the bottom fraction in the absence of membrane from that in the presence of membranes. Targeting to ER is shown by grey bars and targeting to mitochondria is shown by white bars. Error bars represent standard deviation (n=3).
Targeting Cb5 isoforms to ER and mitochondrial membranes
Significant published evidence suggests that the targeting specificity of tail-anchor proteins can be determined by sequences within the CTS, a short sequence of amino acids C-terminal to the hydrophobic TMS [16–18]. To determine what signals within the tail anchor of Cb5A and Cb5B are required to target them specifically to ER, truncation mutants were created in which the CTS was removed (Cb5AΔ and Cb5BΔ). Targeting of the wild-type proteins and the truncation mutants was compared (Cb5A with Cb5AΔ and Cb5B with Cb5BΔ) after incubating the proteins synthesized in a cell-free lysate with either mitochondria or microsomes from ER. To ensure that targeting was taking place post-translationally and therefore independent of the co-translational signal-recognition-particle-mediated targeting system, ribosomes were removed from the translation reactions by centrifugation before the addition of membranes [29]. Targeting of the proteins to membranes was assayed by pelleting the membranes through a sucrose cushion and analysing the distribution of the proteins between pellet and supernatant fractions. A protein was judged membrane-bound if it was recovered from the membrane pellet at the bottom of the tube.
As shown in Figure 2(A), a significant portion of both Cb5A and Cb5B bound to ER since they were found in the pelleted fraction at the bottom of the step gradients. Cb5AΔ and Cb5BΔ targeted to ER with higher efficiency (Figure 2A), indicating the CTS is not required for targeting to ER. While the wild-type versions of Cb5A and Cb5B did not target efficiently to mitochondria, truncation mutants Cb5AΔ and Cb5BΔ did target significantly to mitochondria (Figure 2B; compare bands indicated with arrowheads). Indeed, when the pelleting data from three independent experiments were analysed quantitatively by recording the radioactivity in the SDS/PAGE gels using a phosphoimager, targeting of Cb5AΔ and Cb5BΔ to mitochondria was increased 7- and 4.5-fold respectively compared with the corresponding wild-type proteins (Figure 2C). Only a small portion of each Cb5 construct pelleted in the absence of membranes (Figures 2A and 2B), indicating that the distribution observed was not due to aggregation of the molecules synthesized in vitro. Further, the proteins bound to membranes were integrated in the bilayer as they were resistant to extraction with sodium carbonate (pH 11.5) (results not shown). Therefore, in the absence of the CTS, the mutant proteins targeted efficiently to both ER and mitochondria.
Specificity of Cb5A targeting to ER membranes
The data presented above demonstrate that in targeting experiments conducted with a single source of membranes (mitochondria or ER), Cb5A targeted specifically to ER, whereas Cb5AΔ targeted to either ER or mitochondria (Figure 2). However, there are multiple target subcellular membranes with vastly different surface areas that tail-anchor proteins might target in vivo. Therefore it is possible that even a modest preference for targeting to one organelle might be sufficient to account for targeting specificity in a cell. To examine targeting preference in more detail, an organelle competition assay was used to measure the relative targeting of Cb5A and Cb5AΔ in reactions containing both ER and mitochondrial membranes. Targeting experiments carried out with mitochondria demonstrated that in our cell-free assay 50 μg (total protein) of mitochondria was saturating for targeting of in vitro synthesized Cb5AΔ. Therefore competitive targeting reactions included 50 μg of mitochondria together with increasing amounts of microsomes (Figure 3A). After incubating ribosome-depleted translation reactions with membranes for 60 min at 30 °C, ER and mitochondria were re-isolated via differential centrifugation through a sucrose cushion and the distribution of the proteins between pellet and supernatant fractions was analysed by SDS/PAGE as described above.
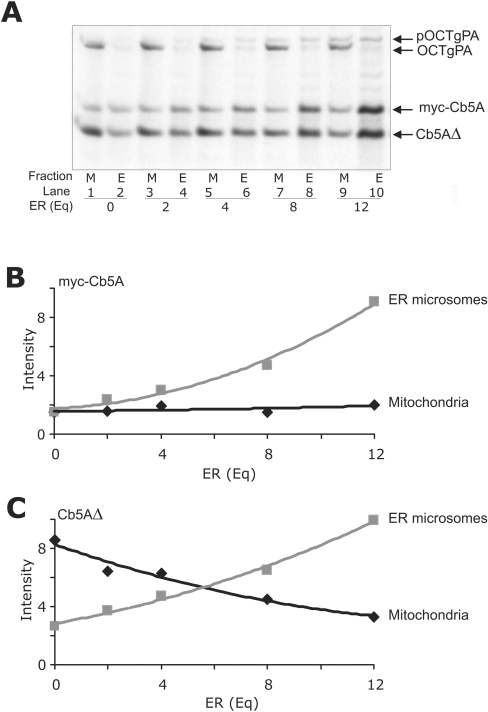
Figure 3. Targeting proteins to ER membranes and mitochondria in the same reactions.
(A) Translation reactions containing Myc–Cb5A, Cb5AΔ and pOCTgPA were added to mitochondrial import reactions containing mitochondria (50 μg of total protein) and equivalents of microsomes as indicated (Eq). After incubation at 30 °C for 60 min, EDTA and potassium acetate were added to final concentrations of 50 and 500 mM respectively, and the ER and mitochondria were separated by differential centrifugation. Equivalent amounts of all fractions were analysed by SDS/PAGE, and the relative intensity of the resulting bands was determined using a phosphoimager (data presented in B and C). Arrows at the side of the panels indicate the migration positions of the specified proteins in SDS/PAGE. pOCTgPA is the precursor containing a mitochondrial targeting signal that is cleaved to generate the mature form of the protein (OCTgPA) following import into mitochondria. (B) Relative amount of Myc–Cb5A co-fractionating with ER (grey squares) and mitochondria (black diamonds). (C) Relative amount Cb5AΔ co-fractionating with ER (grey squares) and mitochondria (black diamonds).
To confirm that the two membranes were efficiently separated after the incubation was completed, mitochondria were labelled during the incubation by import of pOCTgPA into the mitochondrial matrix. pOCTgPA is a previously well-characterized fusion protein composed of the mitochondrial import signal from pOCT (preornithine carbamyl transferase) fused to the N-terminus of the gPA (IgG-binding domain of Staphylococcus aureus protein A) serving as a passive passenger protein [25]. Import of this fusion protein into mitochondria is accompanied by cleavage of the mitochondrial presequence to generate OCTgPA and also demonstrates that the mitochondria in the reactions are import competent ([11] and Figure 3). Separation of OCTgPA from fractions containing ER demonstrates the efficient isolation of mitochondria ([11] and Figure 3).
To compare directly the binding of wild-type and CTS-truncated versions of Cb5A and Cb5B to membranes, sets of proteins were assayed in the same reactions. A Myc epitope tag was added to the N-terminus of wild-type Cb5A and Cb5B to generate sufficient size difference between the full-length and CTS-deleted proteins to separate them cleanly by SDS/PAGE. Addition of the Myc epitope tag at the N-terminus had no effect on binding to membranes or liposomes (results not shown). Quantification of Myc–Cb5A targeting (as described above; Figure 2) revealed that this protein did not target to mitochondria as indicated by the small amount of protein in the mitochondrial pellets (Figure 3A, odd-numbered lanes, and quantified in Figure 3B). However, as the amount of ER was increased in the targeting reactions, the amount of Myc–Cb5A in the ER fraction increased, indicating that Myc–CB5A targets selectively to the ER (Figure 3). As expected from the data in Figure 2, Cb5AΔ targets to mitochondria in the absence of ER (Figure 3A, lanes 1 and 2). However, as the concentration of ER was increased in the targeting reactions, the amount of targeting of Cb5AΔ to ER also increased (Figures 3A and 3C) such that when 12 Eq of ER were added more than half of the Cb5AΔ targeted to ER (lanes 9 and 10). These data for the targeting specificity of Cb5AΔ (Figure 3C) suggest that removal of the CTS from Cb5A results in indiscriminate targeting to the organelle with the highest local concentration. Thus the CTS prevents spontaneous insertion into membranes. In the absence of spontaneous insertion, wild-type tail-anchored proteins undergo specific targeting presumably due to recognition of the tail-anchor sequences by insertion machinery at the appropriate organelle(s) [34–36].
Spontaneous insertion of Cb5AΔ and Cb5BΔ into liposomes
The results of the organelle-targeting experiments described above (Figures 2 and 3) demonstrate that removal of the CTS from Cb5A or Cb5B makes the targeting of these proteins to membranes promiscuous. If targeting specificity in vitro has been lost, then it is possible that these proteins (Cb5AΔ and Cb5BΔ) will now bind to lipid vesicles with a lipid composition similar to that of mitochondria [34].
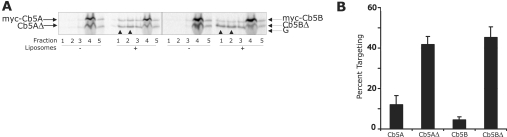
To compare directly the binding of wild-type and CTS-truncated versions of Cb5A and Cb5B to liposomes, sets of proteins were assayed in the same reactions as above. To measure binding of Cb5 proteins to liposomes of mitochondria-like lipid composition, translation reactions containing the proteins were incubated with liposomes and then the liposomes were isolated by flotation on sucrose gradients (Figure 4A, fractions 1 and 2). Unbound proteins remained in the load fraction (fraction 4), while the small amount of protein that aggregated pelleted to the bottom of the gradient (fraction 5). As expected, Myc-tagged Cb5A and Cb5B did not bind efficiently to liposomes (Figure 4A, arrowheads, and quantified in Figure 4B). In contrast, a significant proportion of Cb5AΔ and Cb5BΔ molecules were recovered from fractions 1 and 2, indicating that they bound directly to lipid bilayers (Figure 4A, arrowheads, and quantified in Figure 4B). Note that the globin in the lysate distorts adjacent bands in the gel, limiting the accuracy of quantification. However, because the globin is present only in the load fractions (fraction 4) it actually leads to an underestimation of the extent of liposome binding. Taken together, the organelle and liposome targeting results presented in Figures 2–4 indicate that removal of the hydrophilic CTS sequence allows spontaneous insertion of the Cb5A and Cb5B TMS into any membrane (ER, mitochondria or liposomes) presumably because it lowers the thermodynamic barrier for insertion into lipids.
Figure 4. The Cb5A and Cb5B CTSs inhibit binding to liposomes.
(A) After incubation at 24 °C for 60 min, ribosomes were removed from the translation reactions by centrifugation at 200000 g for 10 min. Phospholipid vesicles were added to translation reactions and, after incubation at 30 °C for 60 min, sucrose was added to a final concentration of 1.6 M. The samples were then transferred to centrifuge tubes and sucrose gradient buffers (0.8 M and 0.25 M sucrose steps) were sequentially layered on top of the sample. After centrifugation for 18 h at 100000 g, gradients were fractionated from the top into five fractions of equal volume with the solubilized pellet as the bottom fraction. Equivalent amounts of all fractions were analysed by SDS/PAGE. Fractions 1 and 2 contain liposomes and liposome-bound proteins (arrowheads). Fraction 3 contains a portion of the sucrose gradient. Fraction 4 is the load fraction and contains untargeted proteins, while fraction 5 contains protein aggregates. The migration positions in SDS/PAGE of each protein examined are indicated by arrows at the sides of the panels. ‘G’ indicates the migration position of rabbit globin in the reticulocyte lysate that distorts the migration of adjacent bands. (B) The percentage of the total protein in fractions 1 and 2 in (A) was determined using a phosphoimager. The percentage targeting reported is corrected for background by subtracting the percentage of protein in fractions 1 and 2 in the absence of liposomes from that in the presence of liposomes. Error bars represent standard deviation from the mean (n=3).
Insertion of Cb5A into ER is protein-mediated
Proteolysed membranes were used to determine if insertion into cellular membranes of the various tail-anchor proteins is dependent on membrane-bound proteins. Trypsin was used to remove cytoplasmic domains from microsomal proteins (including proteins putatively involved in tail-anchor targeting) prior to use of the microsomes in post-translational targeting reactions. RCb5 targeting to microsomes does not rely on a protein receptor [24,34]; therefore this protein served as an appropriate control for membrane integrity. At high concentrations of trypsin microsomes break down, presumably because the proteolytic fragments of transmembrane proteins are more compatible with micellar structures [34]. To ensure that proteolysis was limited to proteins exposed on the microsome surface, degradation of Sec61α compared with calreticulin was monitored by immunobloting. The incubation conditions used (20 μg/ml trypsin, 0 °C for 1 h) resulted in the complete digestion of the cytoplasmic domains of the multi-spanning membrane protein Sec61α, while the luminal protein calreticulin remained intact (results not shown). To measure protein binding to trypsintreated microsomes, the Cb5 proteins were incubated with trypsinized microsomes and then membrane-bound proteins were separated from the reaction by flotation of the microsomes in sucrose gradients. We used separation reactions similar to those described above for liposomes (Figure 4) because trypsin digestion reduces the density of the microsomes by removing ribosomes as well as exposed proteins. As shown in Figure 5(A), RCb5 bound with equal efficiency to both mock-treated (M) and trypsin-treated (T) microsomes, consistent with spontaneous insertion of this protein into bilayers. In contrast with the results obtained for RCb5, incubation of microsomes with trypsin resulted in a significant decrease in targeting of Cb5A compared with targeting to mock-treated microsomes. Consistent with our working hypothesis, targeting of Cb5AΔ to trypsin-digested membranes was not impaired. Together, these results suggest that Cb5A binding to membranes is mediated by a trypsin-sensitive (protein) insertion machinery and that this machinery is not required for binding of Cb5AΔ. Indeed, the latter result was expected since Cb5AΔ inserted into liposomes (Figure 4). The slight decrease in the Cb5AΔ targeting to trypsin-treated microsomes (Figure 5A, compare bands in fractions 1 and 2 for targeting to mock-treated and trypsin-treated microsomes) suggests that while Cb5AΔ undergoes spontaneous insertion it may also make use of the trypsin-sensitive insertion machinery.
Figure 5. Protease sensitivity of targeting of Cb5A and Cb5AΔ to membranes.
(A) Translation reactions containing RCb5, Cb5A or Cb5AΔ were incubated either in the absence of microsomes (–), with mock-treated microsomes (M) or with trypsin-treated microsomes (T). Trypsin-treated microsomes were treated with 20 μg/ml trypsin at 4 °C. After 30 min, PMSF was added to inactivate the protease. Microsomes were washed twice and collected by centrifugation. Mock-treated microsomes were processed the same way as the trypsin-treated microsomes but without addition of trypsin. The indicated membranes were added to translation reactions and after incubation the targeting reactions were then mixed with 2.5 M sucrose gradient buffer to a final sucrose concentration of 1.6 M. Sucrose gradient buffers of 1.3 and 0.8 M sucrose were then layered successively on top of the sample. After centrifugation for 18 h at 440000 g, gradients were fractionated from the top into five fractions of equal volume with the solubilized pellet as the bottom fraction. Equivalent amounts of all fractions were analysed by SDS/PAGE and the relative intensity of the resulting bands was determined using a phosphoimager. Fractions 1 and 2 contain microsomes and microsome-bound proteins (arrowheads) and were used to determine the percentage of microsome-bound protein. Fraction 3 contains a portion of the sucrose gradient. Fraction 4 is the load fraction and contains untargeted proteins, while fraction 5 contains protein aggregates. The migration positions in SDS/PAGE of each protein examined are indicated by arrows at the sides of the panels. (B) Mitochondria were incubated with 0, 5 or 20 μg/ml trypsin (as indicated) at 4 °C and processed as above. Trypsin-treated mitochondria were incubated with translation reactions containing pOCTgPA, RCb5, Cb5AΔ, Cb5AΔ/Bcl or Bcl-2. Targeted molecules were recovered with membrane pellets after centrifugation and analysed by SDS/PAGE. The major band in the pOCTgPA-containing reactions (0 trypsin) is the processed mature OCTgPA protein.
To determine if targeting of Cb5AΔ to mitochondria is also independent of a specific receptor, we exposed mitochondria to different concentrations of trypsin before using them to assay post-translational targeting compared with controls. As expected, targeting of RCb5 to mitochondria does not rely on a membrane receptor [24], whereas the specific receptor-mediated targeting and processing of the other control protein, pOCTgPA, was completely abolished by pre-incubation of mitochondria with 20 μg/ml trypsin (Figure 5B). Thus the targeting of the control proteins to mitochondria monitored well the effects of trypsin. Figure 5(B) shows also that the binding of Cb5AΔ was similar to RCb5 in that it was largely unaffected by treatment of mitochondria with 20 μg/ml of trypsin. The dramatic difference in pOCTgPA targeting compared with Cb5AΔ targeting argues against the involvement of a trypsin-sensitive receptor for targeting of Cb5AΔ to mitochondria. Overall, these results suggest that in vitro the mechanisms of RCb5 and Cb5AΔ binding to membranes are similar and that this mechanism is very different from that of wild-type Cb5A and Cb5B.
Determinants of specificity within the Cb5A CTS
To determine what properties within the Cb5A CTS are important for inhibiting targeting to mitochondria, a series of Cb5A mutants was examined using a cell-free assay containing mitochondria, ER or liposomes (Figure 6). To test the generality of the phenomenon the CTS of Cb5B and a mutant version of the Cb5A CTS (Figure 1) were also examined. The CTS of Cb5B (RLYTKST) contains three fewer charged amino acid residues than the CTS of Cb5A (RHFTKKE). Similarly, Cb5AΔ/mA (RHFTGGE) contains two fewer positive charges. When the CTS of Cb5A was replaced with that of Cb5B, RHFTGGE targeting to both liposomes and mitochondria was reduced compared with Cb5AΔ. However, targeting of Cb5AΔ/B and Cb5AΔ/mA was not entirely similar to Cb5A (Figure 6, compare with Cb5A targeting in Figures 2C and 4B). This result is consistent with previous findings that targeting specificity is a function of the CTS in the context of the appropriate TMS [11]. In addition, the hypothesis that aberrant targeting of Cb5AΔ to mitochondria is the result of spontaneous insertion is supported by the low efficiency of Cb5A/B and Cb5AΔ/mA targeting to both mitochondria and liposomes compared with ER.
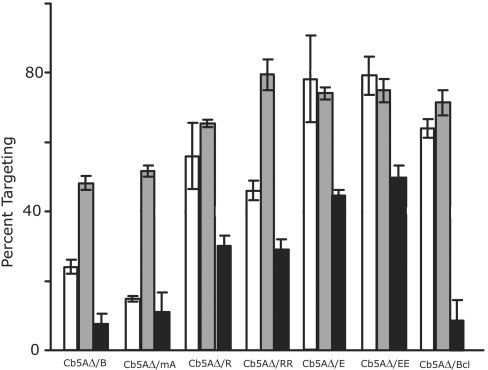
Figure 6. Effect of mutations in the CTS of Cb5A on targeting to ER, mitochondria and liposomes.
Translation reactions containing Cb5AΔ/B, Cb5AΔ/mA, Cb5AΔ/R, Cb5AΔ/RR, Cb5AΔ/E, Cb5AΔ/EE or Cb5AΔ/Bcl were incubated in the presence or absence of ER membranes, mitochondria or liposomes as described in the legends of Figures 2 and 4. The percentage targeting reported is corrected for background by subtracting the percentage of protein in the bottom fraction in the absence of membrane from that in the presence of membranes. Targeting to mitochondria is shown by white bars, targeting to ER is shown by grey bars and targeting to liposomes is shown by black bars. Error bars represent standard deviation from the mean (n=3).
To address the possibility that charges in the CTS of Cb5A are sufficient to provide a thermodynamic barrier to insertion of the hydrophobic tail-anchor TMS into the bilayer, either an arginine or glutamic acid residue was added to the C-terminus of Cb5AΔ, generating Cb5AΔ/R and Cb5AΔ/E respectively. Insertion of Cb5AΔ/R and Cb5AΔ/E into liposomes was then assayed as described above. As shown in Figure 6, Cb5AΔ/E targeted to liposomes with the same efficiency as Cb5AΔ, while the efficiency of liposome binding by Cb5AΔ/R was decreased slightly. The addition of two rather than one arginine or glutamic acid residues to the Cb5AΔ mutant did not further alter liposome targeting efficiency. Therefore addition of one or two charged residues to the TMS of Cb5A does not prevent spontaneous insertion into membranes. Moreover, binding to membranes does not appear to be mediated by electrostatic interactions within the CTS, since mutant Cb5A proteins with more negatively charged CTSs (expected to be repelled from the membrane) targeted more efficiently than those with one or two positively charged residues at their C-termini (Figure 6). Thus the inclusion of two charges does not generate a thermodynamic barrier sufficient to inhibit membrane binding by these tail-anchor proteins and Cb5AΔ/RR and Cb5A/EE target efficiently to both ER and mitochondria. However, targeting of Cb5AΔ/RR to the ER was almost twice as efficient as targeting of this mutant to mitochondria. This suggests that charged amino acids may serve as potential determinants of selectivity. For the above Cb5A mutants a correlation exists between efficient insertion into liposomes and loss of organelle specificity. However, Cb5A/Bcl targets to both ER and mitochondria efficiently, while liposome binding is minimal (Figure 6).
The anti-apoptosis, tail-anchored protein Bcl-2 targets to and functions at both the ER and mitochondria membranes in vitro and in vivo [25,37]. Chemical labelling of the endogenous cysteine in the TMS of Bcl-2 has been used to demonstrate that the tail-anchor of Bcl-2 inserts similarly into the lipid bilayers of both ER and mitochondria in vitro and in cells expressing Bcl-2 [5]. The three-amino-acid CTS of Bcl-2 (SHK; refer to Figure 1) is hydrophilic, suggesting that a protein receptor may be required to allow the CTS to move to the luminal side of the membrane. It has been demonstrated that Bcl-2 targets with reduced efficiency to mitochondria pretreated with trypsin [23]. Addition of the Bcl-2 CTS to Cb5AΔ resulted in efficient targeting to both mitochondrial and ER membranes; however, spontaneous insertion of the mutant Cb5A protein (Cb5AΔ/Bcl) into liposomes was minimal (Figure 6). Targeting to trypsin-treated mitochondria was conducted to determine if addition of the Bcl-2 CTS to CB5AΔ resulted in protein-dependent insertion into mitochondria. Although less sensitive to trypsin treatment than pOCT, the targeting of both Bcl-2 and Cb5AΔ/Bcl is disrupted by trypsin treatment (Figure 5B). Therefore addition of the Bcl-2 CTS prevents spontaneous insertion into liposomes and thereby facilitates protein-mediated insertion into its authentic target membranes (ER and mitochondria). Together with data in Figure 5 demonstrating that Cb5A binding to ER membranes depends on a trypsin-sensitive ER protein, our results suggest that when spontaneous insertion into membranes is prevented, then recognition and insertion of the tail anchor into the correct target membrane is mediated by a membrane-bound accessory factor. For the Cb5 proteins analysed here, the CTS is required to prevent spontaneous insertion and when removed results in efficient but non-specific targeting to ER, mitochondria and liposomes.
DISCUSSION
Many tail-anchored proteins are engaged in cellular processes that require localization to the correct organelle. For some tail-anchored proteins, such as the proteins involved in vesicle budding and fusion (VAMPs and syntaxins), unique localization is an inherent component of function [4]. A few other tail-anchored proteins, such as Bcl-2, function at both ER and mitochondria, but, at least in the case of Bcl-2, have slightly different functions at each location [34,37]. In plants, tung tree Cb5A and Cb5B are localized to and function specifically at the ER, while the equivalent biochemical activity is provided at mitochondria by another distinct Cb5 isoform (Cb5D) that is targeted exclusively to this organelle [11]. Examination of the determinants for organelle-specific targeting of Cb5A, Cb5B and Cb5D in plant cells [11] or mitochondrial-localized Cb5 isoforms in mammalian cells [16] has been informative. However, the molecular mechanism of ER-specific targeting can be better analysed using a cell-free system. Unfortunately, previous examinations of the mechanism of targeting for Cb5 proteins have been hampered by the promiscuous insertion of RCb5 in vitro.
Studies of rabbit and rat isoforms of Cb5 in vitro have shown that promiscuous targeting can result in a hairpin membrane topology [24,38,39]. In the hairpin topology both the N- and C-termini are exposed to the cytosol; therefore the charged CTS does not provide a thermodynamic barrier to spontaneous insertion. Protease protection assays carried out in the present study demonstrated that the bulk of both Cb5A and Cb5AΔ were protease-sensitive when membrane-bound (results not shown). In addition, resistance to carbonate extraction indicated that Cb5A and Cb5B are integral membrane proteins [11]. Nevertheless, our results do not reveal whether the tail anchor actually spans the bilayer. However, our emphasis is on determinants of targeting specificity and since the transmembrane status of either tung Cb5 in cells has not been determined we have not investigated this issue further.
That the plant proteins tung Cb5A and Cb5B retain ER-specific targeting in a mammalian cell-free protein-targeting assay (Figures 2 and 4) suggests that the targeting, signal recognition, membrane-binding and insertion mechanisms are well conserved among evolutionarily diverse organisms. In contrast with expectations based on previous analyses of other tail anchors, changes in the length of the hydrophobic region in the tail anchor examined here are not required to differentiate insertion into ER from insertion into mitochondrial membranes. In the present study, increased targeting to liposomes correlates with increased targeting to mitochondria with the exception of Cb5AΔ/Bcl. That Cb5AΔ/Bcl targets to both ER and mitochondria but not to liposomes or trypsin-treated mitochondria mirrors the targeting specificity of wild-type Bcl-2 [23,34]. Both of these results can be rationalized by the targeting mechanism elucidated here in which a major role of the CTS is to prevent spontaneous insertion into bilayer membranes. However, our results suggest that the Bcl-2 CTS promotes recognition of the Cb5A tail anchor by protein(s) at the mitochondria. The eventual identification of consensus sequences for organelle-specific targeting signals will require: (i) the identification of a much larger number of tail-anchor proteins that are targeted to specific organelles and (ii) largescale mutagenesis of putative targeting signals to assess more accurately the features responsible for targeting a specific subset of tail-anchor sequences.
It was previously reported that Cb5AΔ is localized to the ER in tobacco suspension-cultured cells [11]. This result is consistent with our findings, since we demonstrate that Cb5AΔ in vitro will spontaneously insert into the organelle with the highest local concentration (Figure 4). The ER is by far the most abundant membrane surface in cultured tobacco cells and so it would be expected that most of the Cb5AΔ would localize to this compartment in vivo [11]. In cells, tail-anchor protein translation could be localized such that the protein is synthesized by ribosomes on the ER membrane [40,41]. ER-localized translation would favour spontaneous insertion of Cb5AΔ into ER. Furthermore, in a microscopic examination one also cannot rule out the possibilities that small amounts of Cb5AΔ mislocalized to other organelles were obscured in images by the abundance of Cb5 staining at the ER or that mis-targeted Cb5AΔ protein was rapidly degraded. Unfortunately, the low transfection efficiency of plant cells (less than 1% by biolistic bombardment) precludes the analysis of rates of turnover necessary to test the latter hypothesis. Cell-free targeting assays are well suited to analysis of targeting specificity and efficiency as confounding events in cells such as differences in the rate of turnover of appropriately and mis-targeted proteins are avoided. Furthermore, the amount of each membrane system can be controlled. In our system, we used mitochondria as a non-specific membrane because mitochondria and ER can be efficiently separated and positive controls for import (e.g. pOCTgPA) are readily available. However, our results with liposomes suggest that the results would extend similarly to other off-target organelles. Finally, it is only possible to unambiguously demonstrate post-translational targeting using a cell-free system. For these reasons cell-free targeting reactions are a useful experimental system in which to directly study molecular mechanism(s) involved that confer targeting specificity and efficiency in cis (the present study) or in trans [42,42a].
The targeting signals within the tail-anchor sequences analysed previously all function in cell-free assays as positive signals that direct tail-anchored proteins to a specific membrane (e.g. Figure 7A). Perhaps the best characterized of these signals are those that direct proteins to mitochondria and include essential positive charges that flank an amphipathic TMS [8,16,18]. The essential characteristic identified for a positive signal is that removal disrupts recognition of the tail anchor by the organelle-specific targeting machinery (in this example mitochondria) resulting in decreased targeting to mitochondria and cytoplasmic or ER (mis)localization [9,17]. The observation that some of these proteins localized to ER led to the proposal that for many tail-anchored proteins targeting to the ER occurs by default. Consistent with these previously published results, our data indicate that the ER will tolerate variation in the sequence of the CTS as none of the sequences examined in the context of Cb5A inhibited targeting of this protein to ER (Figure 6). In experiments using transfected cells, the accumulation of Cb5AΔ and Cb5BΔ in the ER may be due to either a specific targeting reaction to ER or spontaneous insertion into lipid bilayers. By using a cell-free assay containing liposomes or microsomes digested with trypsin, we demonstrated that the latter is the more likely mechanism. In this study, the extent of organelle specificity conferred by the CTS of Cb5A is not absolute. It is probable that in addition to the CTS: cytoplasmic factors such as signal recognition particle, targeting signals, membrane-bound proteins and membrane composition all contribute to organelle-specific tail-anchored protein targeting [24,36,40,43,44]. Overall, our results demonstrate a novel mechanism by which targeting specificity for tail-anchored proteins can be achieved. The CTSs of Cb5A and Cb5B prevent spontaneous insertion into a membrane bilayer; therefore translocation of these sequences across the ER membrane is probably protein-mediated (Figure 7B). Evidence from studies using yeast mutants indicates that this machinery for tail-anchored proteins is distinct from the translocon required to insert other membrane proteins into the ER bilayer [36,45]. However, in mammalian cell-free systems, tail-anchored proteins have been shown to cross-link to signal recognition particle and translocon components in vitro [40,43]. Although we did not examine the potential importance of cytoplasmic components in the reticulocyte lysate (such as signal recognition particle), our data with protease-treated microsomal membranes support a role for membrane proteins in the targeting and assembly of tail-anchor proteins into the ER. Furthermore, for some tail-anchor proteins, a lack of targeting information in the protein could be compensated for by targeting the mRNA. Localizing the mRNA to the ER membrane may restrict translation to ER-bound ribosomes thereby targeting an otherwise promiscuous tail-anchor protein in a highly selective manner. In this situation, integration into the membrane may be facilitated by interactions of the tail anchor with the conventional translocation machinery in the ER [40,43]. Little is known about the nature of the tail-anchored protein translocation machinery; it was, therefore, omitted from the diagram in Figure 7. Clearly, identification of the machinery, determination of whether a common machinery is used for all tail-anchor proteins targeted to the ER and how it mediates the translocation of a charged CTS sequence into the ER lumen will all be critical to understanding how membrane insertion and assembly of proteins with tail anchors is regulated.
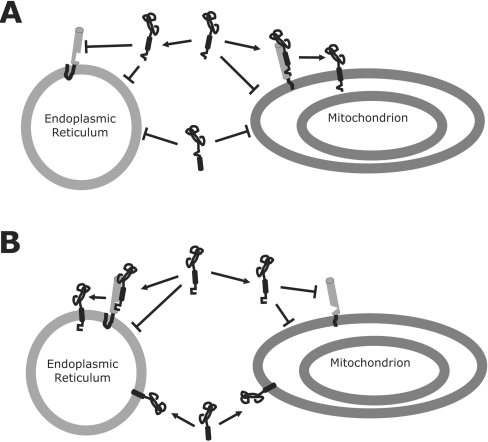
Figure 7. CTSs are involved in targeting tail-anchored proteins to specific organelles.
Recognition of signals by the tail-anchor protein receptor is represented by pairing of squares or triangles on the corresponding receptor and tail-anchor protein. (A) Positive signals promote targeting of a nascent tail-anchored protein to the correct organelle membrane (e.g. mitochondrion membrane) by interacting with a membrane-bound receptor [and/or potentially by recognizing specific lipid composition (not shown)]. When a positive signal is removed the nascent tail-anchored protein remains cytoplasmic. (B) The correct sorting of wild-type Cb5A to the ER is protein-mediated. The CTS of Cb5A (or Cb5B) prevents targeting to the wrong organelle membrane by inhibiting spontaneous insertion into non-target membranes (e.g. mitochondria). However, when the CTS is removed targeting of nascent Cb5A becomes promiscuous. The symbolic representation of tail-anchor recognition by the receptor is illustrative and not intended to imply that the CTS is solely involved, elements within the TMS and NTS are also involved in targeting specificity. Furthermore, cytoplasmic factors may also be involved but are not shown.
Online data
Acknowledgments
This work was supported by grants from the Canadian Institute of Heath Research (FRN 10490) to D.W.A., the United States Department of Agriculture (Current Research Information System project number 6435-41000-083-00D) to J.M.D., and the Natural Sciences and Engineering Research Council (no. 217291) to R.T.M. D.W.A. holds a Canada Research Chair in Membrane Biogenesis, M.P.A.H. is supported by a Natural Sciences and Engineering Research Council Graduate Scholarship, and R.T.M. is a recipient of an Ontario Premier's Research in Excellence Award.
References
- 1.High S., Abell B. M. Tail-anchored protein biosynthesis at the endoplasmic reticulum: the same but different. Biochem. Soc. Trans. 2004;32:659–662. doi: 10.1042/BST0320659. [DOI] [PubMed] [Google Scholar]
- 2.Wattenberg B., Lithgow T. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic. 2001;2:66–71. doi: 10.1034/j.1600-0854.2001.20108.x. [DOI] [PubMed] [Google Scholar]
- 3.Korsmeyer S. J., Wei M. C., Saito M., Weiler S., Oh K. J., Schlesinger P. H. Pro-apoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death. Differ. 2000;7:1166–1173. doi: 10.1038/sj.cdd.4400783. [DOI] [PubMed] [Google Scholar]
- 4.Burri L., Lithgow T. A complete set of SNAREs in yeast. Traffic. 2004;5:45–52. doi: 10.1046/j.1600-0854.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- 5.Kim P. K., Annis M. G., Dlugosz P. J., Leber B., Andrews D. W. During apoptosis bcl-2 changes membrane topology at both the endoplasmic reticulum and mitochondria. Mol. Cell. 2004;14:523–529. doi: 10.1016/s1097-2765(04)00263-1. [DOI] [PubMed] [Google Scholar]
- 6.Annis M. G., Soucie E. L., Dlugosz P. J., Cruz-Aguado J. A., Penn L. Z., Leber B., Andrews D. W. Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. EMBO J. 2005;24:2096–2103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullen R. T., Trelease R. N. The sorting signals for peroxisomal membrane-bound ascorbate peroxidase are within its C-terminal tail. J. Biol. Chem. 2000;275:16337–16344. doi: 10.1074/jbc.M001266200. [DOI] [PubMed] [Google Scholar]
- 8.Beilharz T., Egan B., Silver P. A., Hofmann K., Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- 9.Mihara K. Targeting and insertion of nuclear-encoded preproteins into the mitochondrial outer membrane. Bioessays. 2000;22:364–371. doi: 10.1002/(SICI)1521-1878(200004)22:4<364::AID-BIES6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 10.Walter P., Johnson A. E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 11.Hwang Y. T., Pelitire S. M., Henderson M. P., Andrews D. W., Dyer J. M., Mullen R. T. Novel targeting signals mediate the sorting of different isoforms of the tail-anchored membrane protein cytochrome b5 to either endoplasmic reticulum or mitochondria. Plant Cell. 2004;16:3002–3019. doi: 10.1105/tpc.104.026039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janiak F., Leber B., Andrews D. W. Assembly of Bcl-2 into microsomal and outer mitochondrial membranes. J. Biol. Chem. 1994;269:9842–9849. [PubMed] [Google Scholar]
- 13.Kaufmann T., Schlipf S., Sanz J., Neubert K., Stein R., Borner C. Characterization of the signal that directs Bcl-x(L), but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 2003;160:53–64. doi: 10.1083/jcb.200210084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim P. K., Hollerbach C., Trimble W. S., Leber B., Andrews D. W. Identification of the endoplasmic reticulum targeting signal in vesicle-associated membrane proteins. J. Biol. Chem. 1999;274:36876–36882. doi: 10.1074/jbc.274.52.36876. [DOI] [PubMed] [Google Scholar]
- 15.Suga K., Yamamori T., Akagawa K. Identification of the carboxyl-terminal membrane-anchoring region of HPC-1/syntaxin 1A with the substituted-cysteine-accessibility method and monoclonal antibodies. J. Biochem. (Tokyo) 2003;133:325–334. doi: 10.1093/jb/mvg044. [DOI] [PubMed] [Google Scholar]
- 16.Borgese N., Gazzoni I., Barberi M., Colombo S., Pedrazzini E. Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol. Biol. Cell. 2001;12:2482–2496. doi: 10.1091/mbc.12.8.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horie C., Suzuki H., Sakaguchi M., Mihara K. Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol. Biol. Cell. 2002;13:1615–1625. doi: 10.1091/mbc.01-12-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isenmann S., Khew-Goodall Y., Gamble J., Vadas M., Wattenberg B. W. A splice-isoform of vesicle-associated membrane protein-1 (VAMP-1) contains a mitochondrial targeting signal. Mol. Biol. Cell. 1998;9:1649–1660. doi: 10.1091/mbc.9.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vergeres G., Ramsden J., Waskell L. The carboxyl terminus of the membrane-binding domain of cytochrome b5 spans the bilayer of the endoplasmic reticulum. J. Biol. Chem. 1995;270:3414–3422. doi: 10.1074/jbc.270.7.3414. [DOI] [PubMed] [Google Scholar]
- 20.Borgese N., Colombo S., Pedrazzini E. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J. Cell Biol. 2003;161:1013–1019. doi: 10.1083/jcb.200303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Millar D. G., Shore G. C. Signal anchor sequence insertion into the outer mitochondrial membrane. Comparison with porin and the matrix protein targeting pathway. J. Biol. Chem. 1996;271:25823–25829. doi: 10.1074/jbc.271.42.25823. [DOI] [PubMed] [Google Scholar]
- 22.Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., Endo T., Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- 23.Motz C., Martin H., Krimmer T., Rassow J. Bcl-2 and porin follow different pathways of TOM-dependent insertion into the mitochondrial outer membrane. J. Mol. Biol. 2002;323:729–738. doi: 10.1016/s0022-2836(02)00995-6. [DOI] [PubMed] [Google Scholar]
- 24.Brambillasca S., Yabal M., Soffientini P., Stefanovic S., Makarow M., Hegde R. S., Borgese N. Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J. 2005;24:2533–2542. doi: 10.1038/sj.emboj.7600730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janiak F., Glover J. R., Leber B., Rachubinski R. A., Andrews D. W. Targeting of passenger protein domains to multiple intracellular membranes. Biochem. J. 1994;300:191–199. doi: 10.1042/bj3000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falcone D., Andrews D. W. Both the 5′ untranslated region and the sequences surrounding the start site contribute to efficient initiation of translation in vitro. Mol. Cell. Biol. 1991;11:2656–2664. doi: 10.1128/mcb.11.5.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCartney A. W., Dyer J. M., Dhanoa P. K., Kim P. K., Andrews D. W., McNew J. A., Mullen R. T. Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J. 2004;37:156–173. doi: 10.1111/j.1365-313x.2004.01949.x. [DOI] [PubMed] [Google Scholar]
- 28.Walter P., Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- 29.Andrews D. W., Lauffer L., Walter P., Lingappa V. R. Evidence for a two-step mechanism involved in assembly of functional signal recognition particle receptor. J. Cell Biol. 1989;108:797–810. doi: 10.1083/jcb.108.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenawalt J. W. The isolation of outer and inner mitochondrial membranes. Methods Enzymol. 1974;31:310–323. doi: 10.1016/0076-6879(74)31033-6. [DOI] [PubMed] [Google Scholar]
- 31.Mayer L. D., Hope M. J., Cullis P. R. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim. Biophys. Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 32.Song W., Raden D., Mandon E. C., Gilmore R. Role of Sec61α in the regulated transfer of the ribosome–nascent chain complex from the signal recognition particle to the translocation channel. Cell. 2000;100:333–343. doi: 10.1016/s0092-8674(00)80669-8. [DOI] [PubMed] [Google Scholar]
- 33.Schagger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 34.Kim P. K., Janiak-Spens F., Trimble W. S., Leber B., Andrews D. W. Evidence for multiple mechanisms for membrane binding and integration via carboxyl-terminal insertion sequences. Biochemistry. 1997;36:8873–8882. doi: 10.1021/bi970090t. [DOI] [PubMed] [Google Scholar]
- 35.Lan L., Isenmann S., Wattenberg B. W. Targeting and insertion of C-terminally anchored proteins to the mitochondrial outer membrane is specific and saturable but does not strictly require ATP or molecular chaperones. Biochem. J. 2000;349:611–621. doi: 10.1042/0264-6021:3490611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steel G. J., Brownsword J., Stirling C. J. Tail-anchored protein insertion into yeast ER requires a novel posttranslational mechanism which is independent of the SEC machinery. Biochemistry. 2002;41:11914–11920. doi: 10.1021/bi026105r. [DOI] [PubMed] [Google Scholar]
- 37.Zhu W., Cowie A., Wasfy G. W., Penn L. Z., Leber B., Andrews D. W. Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. EMBO J. 1996;15:4130–4141. [PMC free article] [PubMed] [Google Scholar]
- 38.Enoch H. G., Fleming P. J., Strittmatter P. The binding of cytochrome b5 to phospholipid vesicles and biological membranes. Effect of orientation on intermembrane transfer and digestion by carboxypeptidase Y. J. Biol. Chem. 1979;254:6483–6488. [PubMed] [Google Scholar]
- 39.Vergeres G., Waskell L. Cytochrome b5, its functions, structure and membrane topology. Biochimie. 1995;77:604–620. doi: 10.1016/0300-9084(96)88176-4. [DOI] [PubMed] [Google Scholar]
- 40.Abell B. M., Pool M. R., Schlenker O., Sinning I., High S. Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 2004;23:2755–2764. doi: 10.1038/sj.emboj.7600281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicchitta C. V. A platform for compartmentalized protein synthesis: protein translation and translocation in the ER. Curr. Opin. Cell Biol. 2002;14:412–416. doi: 10.1016/s0955-0674(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 42.Andrews D. Examining protein translocation in cell-free systems and microinjected Xenopus oocytes. BioTechniques. 1989;7:960–962. 964–967. [PubMed] [Google Scholar]
- 42a.Andrews D. Examining protein translocation in cell-free systems and microinjected Xenopus oocytes. BioTechniques. 1989;7:964–967. [PubMed] [Google Scholar]
- 43.Abell B. M., Jung M., Oliver J. D., Knight B. C., Tyedmers J., Zimmermann R., High S. Tail-anchored and signal-anchored proteins utilize overlapping pathways during membrane insertion. J. Biol. Chem. 2003;278:5669–5678. doi: 10.1074/jbc.M209968200. [DOI] [PubMed] [Google Scholar]
- 44.Halbach A., Landgraf C., Lorenzen S., Rosenkranz K., Volkmer-Engert R., Erdmann R., Rottensteiner H. Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J. Cell Sci. 2006;119:2508–2517. doi: 10.1242/jcs.02979. [DOI] [PubMed] [Google Scholar]
- 45.Yabal M., Brambillasca S., Soffientini P., Pedrazzini E., Borgese N., Makarow M. Translocation of the C terminus of a tail-anchored protein across the endoplasmic reticulum membrane in yeast mutants defective in signal peptide-driven translocation. J. Biol. Chem. 2003;278:3489–3496. doi: 10.1074/jbc.M210253200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.