Abstract
Regulated proteolysis of the polyprotein precursor by the NS2B–NS3 protease is required for the propagation of infectious virions. Unless the structural and functional parameters of NS2B–NS3 are precisely determined, an understanding of its functional role and the design of flaviviral inhibitors will be exceedingly difficult. Our objectives were to define the substrate recognition pattern of the NS2B–NS3 protease of West Nile and Dengue virises (WNV and DV respectively). To accomplish our goals, we used an efficient, 96-well plate format, method for the synthesis of 9-mer peptide substrates with the general P4–P3–P2–P1–P1′–P2′–P3′–P4′–Gly structure. The N-terminus and the constant C-terminal Gly of the peptides were tagged with a fluorescent tag and with a biotin tag respectively. The synthesis was followed by the proteolytic cleavage of the synthesized, tagged peptides. Because of the strict requirement for the presence of basic amino acid residues at the P1 and the P2 substrate positions, the analysis of approx. 300 peptide sequences was sufficient for an adequate representation of the cleavage preferences of the WNV and DV proteinases. Our results disclosed the strict substrate specificity of the WNV protease for which the (K/R)(K/R)R↓GG amino acid motifs was optimal. The DV protease was less selective and it tolerated well the presence of a number of amino acid residue types at either the P1′ or the P2′ site, as long as the other position was occupied by a glycine residue. We believe that our data represent a valuable biochemical resource and a solid foundation to support the design of selective substrates and synthetic inhibitors of flaviviral proteinases.
Keywords: Dengue fever virus, flavivirus, West Nile virus, NS2B–NS3, peptide cleavage assay, polyprotein precursor processing
Abbreviations: BOP, benzotriazol-1-yl-oxy-tris-(dimethylamino)-phosphoniumhexafluorophosphate; DV, Dengue virus; FAM, 6(5)-carboxyfluorescein; Fmoc, fluoren-9-ylmethoxycarbonyl; NS2B and NS3, non-structural viral proteins 2B and 3 respectively; NS3hel, the helicase domain of the NS3 protein; NS3pro, the proteinase domain of the NS3 protein; PEG, poly(ethylene glycol); WNV, West Nile virus
INTRODUCTION
WNV (West Nile virus) and DV (Dengue fever virus) are members of the Flaviviridae family. WNV and DV are transmitted to animals, including humans, by mosquito bites. Both WNV and DV have an icosahedral core (30- to 35-nm in size) composed of multiple copies of a 12 kDa capsid protein [1,2]. The capsid encloses a single-stranded RNA with a single reading frame encoding a polypeptide precursor of approx. 3400 amino acid residues [3]. There are three structural proteins [C (capsid), prM (membrane) and E (envelope)] and seven NS (non-structural) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) encoded by the flaviviral genome. Proteases from the host (furin and secretase) and from the virus [NS3pro (NS3 serine proteinase)] are required to process the polyprotein precursor into the individual functional proteins [4–19].
The full length NS3 peptide sequence represents a multifunctional protein [20–23]. The N-terminal 184 amino acid long fragment represents NS3pro. The C-terminal portion of the NS3 protein encodes a nucleotide triphosphatase, an RNA triphosphatase and a helicase. NS3pro is responsible for the cleavage of the capsid C protein and for cleavage at the NS2A/NS2B, NS2B/NS3, NS3/NS4A, NS4A/NS4B (probably) and NS4B/NS5 boundaries [6,24]. As is the case with a number of flaviviruses, the NS2B protein, which is located in the polypeptide precursor upstream of the NS3pro domain, functions as a cofactor and promotes the folding and the functional activity of the NS3 proteolytic enzyme [4,5,9,18,25–27]. The cofactor activity of the 48 amino acid long central portion of the NS2B is roughly equivalent to that of the entire NS2B sequence [26]. Inactivating mutations of the NS3pro cleavage sites in the polyprotein precursor abolished viral infectivity [3,28]. These parameters suggest that NS3pro is a promising drug target for flaviviral inhibitors.
As a step towards inhibitor design, we have mapped the substrate recognition specificity by NS3pro from WNV and DV. Our results disclosed the unexpectedly strict substrate selectivity and specificity of the WNV protease, especially at the P1′ and P2′ sites when compared with that of the DV enzyme, and they provide a starting point for developing novel diagnostic assays and therapeutics directly aimed at a broad range of flaviviruses.
EXPERIMENTAL PROCEDURES
Reagents
All reagents were purchased from Sigma–Aldrich, unless indicated otherwise. All solvents for peptide synthesis were from VWR International. Fmoc (fluoren-9-ylmethoxycarbonyl) amino acids, BOP reagent [benzotriazol-1-yl-oxy-tris-(dimethylamino)-phosphoniumhexafluorophosphate] and biotin resin were purchased from EMD Biosciences. The furin and WNV NS2B–NS3 proteases were purified as described previously [29,30]. A fragment of the WNV strain NY99 cDNA that included the sequence of NS2B–NS3 proteins was kindly provided by Dr Richard Kinney (Centers for Disease Control and Prevention, Fort Collins, CO, U.S.A.). A fragment of the DV serotype 2 cDNA (strain 16681) that included the sequence encoding the NS2B–NS3 proteins was a kind gift of Dr Michael Diamond (Washington University Medical School, St.Louis, MO, U.S.A.). Furin was kindly provided by Dr Iris Lindberg (Louisiana State University, New Orleans, LA, U.S.A.).
The WNV and DV NS2B–NS3 expression constructs
The 5′-GGGGGCGGAGGTAGTGGTGGACAACAGGCTGGAGTATTGTGGGATG-3′ and 5′-TATAGCTTCGCTGACTATGGCCGG-3′ oligonucleotides as direct and reverse primers respectively, and the DV serotype 2 cDNA fragment as a template were used in PCR reactions to generate the NS3 sequence. The 5′-CACCATGTCGGCCGATTTGGAACTGGAGAGAGCAGCCG-3′ (direct primer) and 5′-CTGTTGTCCACCACTACCTCCGCCCCCCAGTGTTTGTTCTTCCTC-3′ (reverse primer) were used to amplify the 48 residue long central region of the NS2B protein. The NS2B part (amino acids 1393–1440) and the NS3pro part (amino acids 1476–1687) were linked by a nona-peptide linker GGGGSGGQQ (in the primers, the linker sequence is underlined). The final 804-bp NS2B-GGGGSGGQQ-NS3 construct was amplified by PCR and its authenticity was confirmed by sequencing. The NS2B–NS3 DV construct was cloned into the pET101 Topo cloning vector (Invitrogen).
The WNV 48 amino acid NS2B sequence was linked via a GGGGSGGGG linker to the WNV NS3pro sequence as described previously [29]. The WNV catalytically inert NS2B–NS3pro H51A mutant was obtained by using a QuikChange® mutagenesis kit (Stratagene) and the oligonucleotides 5′-GTTTTCCACACCCTTTGGGCTACAACAAAAGGAGCCGC-3′ and 5′-GCGGCTCCTTTTGTTGTAGCCCAAAGGGTGTGGAAAAC-3′ as the forward and reverse primers (mutant positions are underlined). The autolytic-site-deficient NS2B–NS3pro K48A mutant construct was prepared with the 5′-CCAGGAGCACCTTGGGCGGGCGGGGGAGGT-3′ and 5′-ACCTCCCCCGCCCGCCCAAGGTGCTCCTGG-3′ forward and reverse primers respectively (mutant nucleotides are underlined). The NS3hel domain sequence (amino acids 1686–2118) was amplified by PCR using the 5′-GCCGGATTCGAACCTGAGATGCTG-3′ and 5′-TCAATGGTGATGGTGATGATGTCCCGAGGCGAAGTCCTTGAACGCC-3′ oligonucleotides as the forward and reverse primers respectively (the sequence encoding the His-tag is underlined) and the WNV cDNA as a template. The NS3hel domain sequence was ligated to the autolytic site-deficient NS2B–NS3pro K48A mutant and to the proteolytically inert NS2B–NS3pro H51A mutant constructs to obtain the full-length NS2B–NS3prohel K48A and the NS2B–NS3pro-hel H51A WNV sequences respectively. The constructs were re-cloned into the pET101 expression vector after confirming their authenticity by sequencing.
Enzyme expression and purification
Competent Escherichia coli BL21 (DE3) Codon Plus cells (Stratagene) were transformed with recombinant pET101 vectors. Transformed cells were grown in 2 litres of Luria–Bertani broth containing 0.1 mg/ml ampicillin at 30 °C. Cultures were induced with 0.6 mM IPTG (isopropyl β-D-thiogalactoside) and growth was continued for an additional 16 h at 18 °C. The cells were then collected by centrifugation (5000 g for 15 min), re-suspended in PBS containing 1 M NaCl and 10 mg/ml lysozyme and disrupted by sonication. The pellet was removed by centrifugation at 20000 g for 30 min. The recombinant DV NS2B–NS3pro and the wild-type and mutant WNV NS2B–NS3pro and NS2B–NS3pro-hel constructs, C-terminally tagged with a hexahistidine tag, were purified from the supernatant fraction using affinity chromatography on a Co2+-chelating Sepharose Fast Flow column [29]. To determine the autolytic cleavage site, the purified individual WNV NS3pro was subjected to N-terminal sequencing at ProSeq.
High-throughput peptide synthesis
High-throughput peptide synthesis was performed in wells of a 96-well flat bottom polypropylene microtitre plate (Evergreen Scientific) in a centrifugal peptide synthesizer as described previously [31,32]. The resin (Nova Tag, Novabiochem/EMD Bioscience) modified with Fmoc-Gly-biotin-PEG [poly(ethylene glycol)] was used for the synthesis of the peptides. Peptides were assembled using an Fmoc chemistry and BOP as a coupling reagent. 4-Methylpiperidine was used instead of piperidine (a regulated substance) for the Fmoc group removal [33]. Following the modification of the N-termini of the peptides with FAM [6(5)-carboxyfluorescein], the peptides were treated with a 4-methylpiperidine solution to remove by-products resulting from the coupling of FAM to free hydroxy groups [34]. A mixture with a 39:61 ratio of the 5- and 6-FAM isomers was used. The peptides were then cleaved from the resin by a trifluoroacetic acid/thioanisol/water/phenol/ethanedithiol mixture [82.5:5:5:5:2.5 (v/v)] [35]. As a result, the prepared peptides were C-terminally and N-terminally tagged with biotin and FAM respectively. The yield of the synthesized individual peptides was normally 1 mg/well.
The purity of the peptides was confirmed by reverse-phase HPLC on a μBondapak C18 column [10 μ, 125 Å (1 Å=0.1 nm), 150×3.9 mm i.d. (internal diameter)] using a gradient of solvent (A) 0.05% trifluoroacetic acid/water and solvent (B) 0.05% trifluoroacetic acid/70% acetonitrile (from 5% to 60% of B in 15 min) on an Agilent 1100 HPLC chromatographer (Palo Alto) and also by MS.
Peptide cleavage assay
Aliquots (3 μg each) of the synthesized peptides were dissolved in 0.3 ml of 10 mM Tris/HCl buffer (pH 8.0), containing 20% glycerol. The 0.1 ml aliquots of the peptide solutions were transferred into the wells of three identical black, flat-bottom 96-well plates. The first plate was used to determine the maximum fluorescence of the peptides. A Tecan fluorescence reader was used for the fluorescence measurements (λex 492 nm and λem 535 nm). For the complete pull-down of the peptides a 30 μl aliquot [0.6% slurry (w/v); 4.5 nmol/mg] of streptavidin-coated magnetic beads (Seradyn) was added to each well of the second plate. After a 15 min incubation at ambient temperature (20 °C), magnetic beads were sedimented by placing the plate into a magnetic particle concentrator Dynal MPC-96S (Invitrogen) for 2 min. The residual fluorescence of the supernatant fractions was measured to determine the non-specific background fluorescence. To induce the exhaustive cleavage of the peptides, a 1 μg aliquot (1 μl; 0.3 μM) of NS2B–NS3pro was added to each well of the third plate. After a 2 h incubation at 37 °C the plates were transferred on ice to block further cleavage. To pull-down the biotin-labelled C-terminal cleavage products and the residual amounts of the intact peptides, a 30 μl aliquot [0.6% slurry (w/v); 4.5 nmol/mg] of streptavidin-coated magnetic beads was added to each well. After a 15 min incubation, magnetic beads were sedimented by placing the plate into a magnetic particle concentrator Dynal MPC-96S (Invitrogen) for 2 min. The fluorescence of the N-terminal, FAM-tagged, cleavage products which were present in the supernatant fraction was measured on a fluorescence reader. The percentage of the peptide proteolysis was calculated using the following equation: C−B/A−B, where A, B and C are the A535 values of the first, second and third plate respectively.
Cleavage of peptides and MS analyses
The peptides (1 μg; approx. 30 μM) were incubated with the NS2B–NS3pro constructs (0.7 μg; 1.25 μM) for 2 h at 37 °C in 20 μl of 10 mM Tris/HCl buffer (pH 8.0), containing 20% glycerol. The molecular mass of the intact peptides and the digest products was determined by MALDI–TOF MS (matrix-assisted laser-desorption ionization–time-of-flight MS) analysis using an Autoflex II mass-spectrometer (Brucker Daltonics).
General methods
All of the general methods used including Western blotting, enzyme kinetics and protease assays with fluorogenic substrates, inhibition assays and related techniques, have been described in our previous publication [29].
RESULTS
NS3 constructs
Previous studies have shown that the presence of the 48 residue central NS2B domain linked to the N-terminus of NS3 significantly enhanced the accumulation of soluble recombinant NS3pro in E. coli [24]. We used a similar approach to express the wild-type and mutant WNV NS2B–NS3pro and the DV NS2B–NS3pro. In the expression constructs, the NS2B sequence was linked with the NS3pro domain via a GGGGSGGGG linker. To improve the crystallization properties of the protein, the linker sequence was insignificantly modified in the DV construct (GGGGSGGQQ). The structure of the WNV and DV constructs and the relative positions of the mutations are shown in Figure 1.
Figure 1. Constructs of the NS2B–NS3 proteinase of WNV and DV.
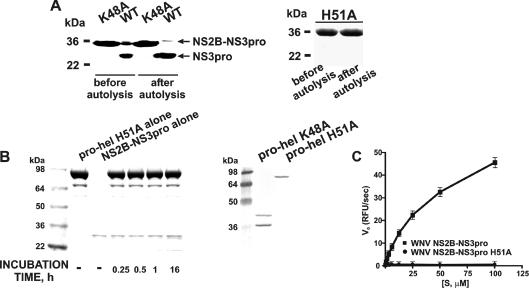
The central portion of NS2B (short NS2B) was linked with the His-tagged NS3pro sequence (the linker sequence is underlined). The WNV NS2B–NS3pro construct autolytically cleaves the G49↓G50 bond. The alanine residue substituted for the essential His51 in the H51A inert mutant. The Ala substituted for Lys48 in the autolytic-site-deficient K48A mutant. The K48A and H51A NS2B–NS3pro constructs were linked to the NS3hel sequence to obtain the full-length NS2B–NS3pro-hel K48A (autolytic-site-deficient) and NS2B–NS3 H51A (proteolytically inert) mutant constructs.
To evaluate the sensitivity of the junction region between the WNV NS3pro-hel domains to the viral protease, we first inactivated the autolytic cleavage site in the NS2B/NS3 boundary [29,36]. We identified the cleavage site sequence by incubating the isolated NS2B–NS3pro construct overnight to induce the autolytic cleavage of the NS2B/NS3 junction (Figure 2). We then determined the N-terminal peptide sequence of the individual NS3pro domain (Figure 1). The N-terminal sequence of the individual WNV NS3pro was determined to be GGGSGG suggesting that in the course of autolysis the NS3 protease activity cleaved, in an unconventional way, the KG↓GGGSGGGG linker sequence (the linker sequence is underlined). We believe that this in cis cleavage may be explained by the loop-like structure and the favourable presentation of the linker to the active site of the protease. Based on these sequence data, we constructed the K48A mutant. The K48A mutant construct included the 48 amino acid residue NS2B sequence and the sequence of the NS3pro domain. The K48A mutation of the C-terminal amino acid residue of the NS2B sequence inactivated the autolytic cleavage site. As a result, the K48A NS2B–NS3pro mutant was determined to be resistant to autoproteolysis (Figure 2). The K48A mutation, however, did not have any significant effect on the catalytic activity of the NS2B–NS3pro construct and the K48A construct was highly efficient in cleaving the t-butoxycarbonylRVRR-7-amino-4-methylcoumarin and pyroglutamic acid-RTKR-7-amino-4-methylcoumarin fluorescent peptide substrates (Table 1).
Figure 2. The properties of the mutant WNV NS2B–NS3 constructs.
(A) The NS2B–NS3pro K48A (left-hand panel) and the NS2B–NS3 H51A (right-hand panel) mutants are resistant to autoproteolysis. The purified NS2B–NS3pro (WT) and the K48A and H51A constructs (before autolysis) were incubated overnight (after autolysis). The samples were separated by gel-electrophoresis and stained with Coomassie Blue. (B) The inert NS2B–NS3pro-hel H51A construct is not cleaved by NS2B–NS3pro in trans, but the NS2B–NS3pro-hel K48A mutant is cleaved by the integral NS3pro in cis. Left-hand panel (Coomassie Blue staining), NS2B–NS3prohel H51A (pro-hel H51A) was incubated with the active WNV NS2B–NS3pro. Right-hand panel, the NS2B–NS3pro-hel K48A and H51A mutants were analysed by Western blotting of the E. coli cell lysate aliquots using the antibody the C-terminal His-tag. The individual, untagged, NS2B–NS3pro moiety is not visible on the blots. (C) NS2B–NS3pro H51A mutant does not cleave pyroglutamic acid-RTKR-7-amino-4-methylcoumarin. RFU, relative fluorescence units; Vo, the initial velocity of substrate hydrolysis; [S], substrate concentration.
Table 1. The wild-type and the K48A mutant WNV NS2B–NS3pro cleave fluorescent peptide substrates.
| Pyroglutamic acid-RTKR-7-amino-4-methylcoumarin | t-Butoxycarbonyl-RVRR-7-amino-4-methylcoumarin | |||||
|---|---|---|---|---|---|---|
| NS2B–NS3pro | Km (μM) | kcat (s−1) | Kcat/Km (M−1·s−1) | Km (μM) | kcat (s−1) | Kcat/Km (M−1·s−1) |
| WNV wild-type | 71±15 | 6.3±0.4 | 88000±12000 | 40±7 | 0.22±0.01 | 5500±500 |
| WNV K48A | 59±10 | 5.3±0.3 | 89300±11000 | 36±6 | 0.18±0.01 | 5000±500 |
We next constructed the WNV NS2B–NS3pro H51A mutant. This construct included the 48 amino acid residue NS2B sequence and the NS3pro sequence. The H51A mutation of the catalytically essential His51 inactivated the protease active site. As a result, this mutant exhibited no proteolytic activity (Figure 2). Lastly and specifically for our follow-up crystallization efforts, we constructed the WNV NS2B–NS3pro-hel H51A mutant. This catalytically inert mutant included the 48 amino acid residue NS2B sequence and the full-length NS3 sequence that represented both the protease (pro) and the helicase (hel) domains. The H51A mutant was devoid of any proteolytic activity and, consequently, was incapable of autolytically cleaving either the NS2B sequence or the NS3pro-hel boundary. In addition, the NS2B–NS3pro-hel H51A WNV mutant protein was totally resistant to the proteolysis in trans by the external, highly active NS2B–NS3pro WNV construct thus suggesting the absence of the accessible cleavage sites to the NS3 proteolytic activity in the full-length NS2B–NS3pro-hel H51A sequence (Figure 2). In contrast, the full-length autolytic-site-deficient NS2B–NS3pro-hel K48A mutant was readily cleaved in cis by the integral NS2B–NS3pro activity and, as a result of self-cleavage, generated two cleavage species of the individual NS3hel domain (Figure 2).
Peptide cleavage screening assay
The main objective of the present study was to define the scope of substrate recognition by WNV NS2B–NS3pro and to compare its recognition patterns with that of the closely related DV type 2 NS2B–NS3pro. Our previous studies suggested that WNV NS2B–NS3pro exhibited a furin-like cleavage preference and that it required the presence of a lysine or arginine residue at the P2 position and an arginine residue at the P1 position to achieve the efficient cleavage of the peptide substrate [29]. Studies in other laboratories have indicated that similar cleavage preferences for basic amino acid residues (arginine/lysine) at both the P1 and P2 positions of the DV NS3 protease activity also exist and that the cleavage motifs have features in common with the physiological cleavage sites in flaviviral polyprotein precursors [37,38]. Consistent with these findings, we biased our library of the 9-mer peptides to the cleavable sequences and we avoided those peptide sequences which we believed would be resistant to NS2B–NS3 proteolysis.
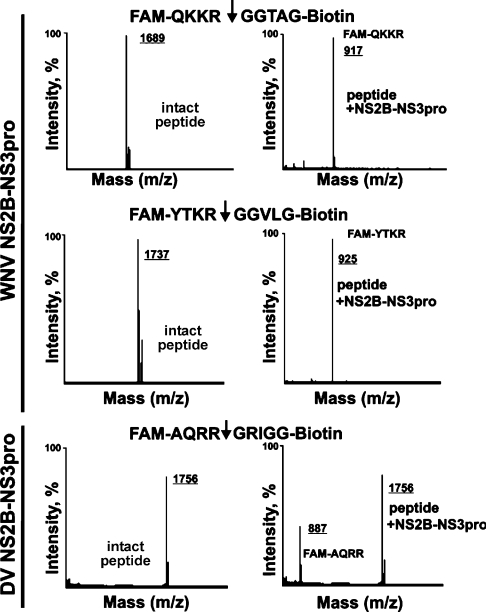
A novel peptide synthesis method that employed a centrifugal, 96-well format, peptide synthesizer was used to synthesize the peptides required for the present study. To facilitate the follow-on cleavage screening assay, the prepared peptides were C-terminally and N-terminally tagged with biotin and FAM respectively, in the course of the synthesis. The high quality of the peptides was confirmed by HPLC (Figure 3) and MS analyses (Figure 4).
Figure 3. Representative example of the reverse-phase HPLC profile of the synthesized peptides.
HPLC confirms the purity of the Fam-FLKRYAEA-Gly-PEG-biotin peptide. Because a 39:61 mixture of the 5- and 6-FAM isomers was used for tagging, two major peptide forms were observed.
Figure 4. MS analysis of the cleavage peptides.
The FAM-Q102KKR↓GGTA109G-biotin and FAM-Y1498TKR↓GGVL1505G-biotin peptides (from the sequence of the WNV capsid C protein and the NS2B/NS3 boundary respectively) were cleaved by WNV NS2B–NS3pro. Similarly, the FAM-A1930QRR↓GRIG1937G-biotin peptide (the potential cleavage site in the central part of DV NS3hel) was cleaved by DV NS2B–NS3pro. The molecular mass of the peptides was determined by MS. There was no difference between the calculated and the estimated molecular mass of the peptides.
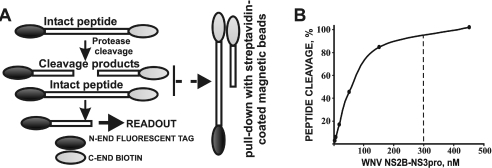
Previous studies have also verified the efficiency of the synthesizer and the synthetic scheme we used and the high quality of the synthesized peptides [31–33,39,40]. As a proof-of-principle, the same approach was used for the synthesis of the peptide sequences, which were used as the cleavage targets of trypsin, chymotrypsin, caspase-3, subtilisin-A, enterokinase and tobacco etch virus protease [41]. These additional data verified that the synthetic method and the follow-on peptide cleavage screening were applicable for the analyses of many endoprotease types instead of for the NS2B–NS3pro alone. We are now confident that the designed peptide synthesis and peptide cleavage assay methods will be widely used by other laboratories interested in using a time-saving, efficient method of rapidly and precisely determining the cleavage preferences of proteolytic enzymes. The peptide synthesis was followed by a 96-well format, peptide cleavage screening assay (Figure 5). To screen the peptides and to distinguish the peptides which are either resistant or are poorly sensitive to NS2B–NS3, we specifically selected the exhaustive proteolysis conditions. Thus 1 μg (approx. 300 nM concentration) of NS2B–NS3pro was sufficient for the exhaustive hydrolysis of a 1 μg aliquot of peptides in our experimental conditions. The dynamic range of the screening method is presented in Figure 5. After an exhaustive proteolysis, both the residual amounts of the intact peptides and the C-terminal cleavage products were pulled-down by streptavidin-coated magnetic beads. The fluorescence of the N-terminal, FAM-tagged products, which were generated by the proteolysis of the FAM-P4-P4′-biotin peptides, was measured. The percentage of the peptide proteolysis was calculated using the equation that is shown in the Experimental procedures section.
Figure 5. Peptide cleavage assay.
(A) The Gly-biotin- and FAM-tagged peptides were incubated with NS2B–NS3pro. After an exhaustive proteolysis, the biotin-labelled cleavage products and the residual intact peptides were pulled-down with streptavidin-coated magnetic beads. The fluorescence of the FAM-tagged cleavage products was then measured. (B) The dynamic range of the cleavage reactions. The FAM-G2522LKR↓GGAK2529-Gly-biotin peptide was cleaved by WNV NS2B–NS3pro for 2 h. The fluorescence of the generated FAM-tagged GLKR was measured. Based on these dynamic range data, a 300 nM concentration of NS2B–NS3pro (marked by a dashed line) was routinely used in the cleavage screening assays.
In addition to the results of HPLC and MS (Figures 3 and 4), the reliability of the synthesis and the accuracy of the assay were further confirmed by the analysis of triplicate and duplicate samples of the multiple peptides. For example, WNV NS2B–NS3pro proteolysis of two batches of the peptide NRKR↓GGPA resulted in 78% and 77% cleavage. Cleavage of the peptide QRRR↓GGTA by WNV NS2B–NS3pro twice resulted in a 58% cleavage. Three individual batches of the peptide AQRR↓GRIG resulted in 3%, 2% and 0% cleavages by WNV NS2B–NS3pro. Similarly, DV NS2B–NS3pro generated 45%, 37% and 36% cleavages of three batches of the peptide AQRR↓GRIG.
The analysis of WNV and DV NS2B–NS3pro
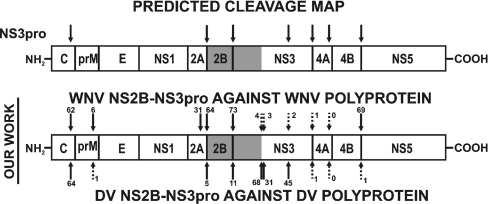
As a starting point for our screening studies, we synthesized the peptides which span the potential NS3pro cleavage sites in the WNV and DV precursor polyprotein (Table 2). The peptide sequences were cleaved by the WNV and DV proteases to confirm their role in flaviviral polyprotein precursor processing. In agreement with multiple previous publications, both proteases efficiently cleaved the peptides derived from the WNV and DV capsid protein C (Q102KKR↓GGTA109 and R97RRR↓SAGM104 respectively), from the NS2A/NS2B junction region (N1367RKR↓GWPA1374 and S1342KKR↓SWPL1349 respectively) and NS2B/NS3 (Y1498TKR↓GGVL1505 and K1472KQR↓AGVL1479 respectively). In addition, WNV NS2B–NS3pro efficiently cleaved the peptide that represented the NS4B/NS5 junction region (G2522LKR↓GGAK2529), but DV NS2B–NS3pro did not cleave the corresponding peptide (N2488TRRGTGN2495) from the NS4B/NS5 junction region of the DV polyprotein. We also determined that the peptides S2117GKRSQIG2124 and A2090GRKSLTL2097 that represented the NS3/NS4A junction regions of the WNV and DV polyproteins respectively, were resistant to the viral proteinases. The peptides E2243KQRSQTD2250 and E2217KQRTPQD2224 derived from the putative cleavage sites of the NS4A/NS4B of the WNV and DV polyproteins respectively, were also resistant to the viral proteinases. We also determined that the peptides R1659KRR↓LTIM1666 and K1674TKR↓YLPA1681 from the putative junction region of the DV NS3 protease-helicase domains were cleaved by DV NS2B–NS3pro. In contrast, the efficiency of the cleavage of the corresponding peptides R1686KKQ↓ITVL1693 and K1700TRK↓ILPQ1707 from the putative junction region of the WNV NS3 protease-helicase domains, was low. The WNV NS2B–NS3pro-hel H51A proteolytically inert mutant was resistant to the proteolysis in trans by the external WNV NS2B–NS3pro activity (Figure 2). In contrast, the proteolytically potent, albeit the autolytic-cleavage-site deficient, WNV NS2B–NS3pro-hel K48A construct was readily cleaved in cis by the integral NS3pro activity, suggesting that the specific presentation of the NS3pro/hel boundary sequence region to the active site of the NS3pro domain allows to overcome the resistance to proteolysis of the suboptimal cleavage sequences.
Table 2. The potential NS2B–NS3pro cleavage sites in the precursor polyprotein.
The sequence of the synthesized and tested peptides which span the potential cleavage sites in the WNV and DV polyproteins is shown. The cleavage sites represent the boundaries between the individual proteins in the polyprotein precursor. The additional, dibasic, potential cleavage sites were identified via the analysis of the polyprotein peptide sequence. The efficiency of the cleavage (%) is shown in parentheses.
| Proteins | WNV | DV |
|---|---|---|
| Capsid C protein | Q102KKR↓GGTA109 (62) | R97RRR↓SAGM104 (64) |
| C/prM | C120AGA↓VTLS127 (1) | T111VMA↓FHLT118 (0) |
| prM | H209SRR↓SRRS216 (6) | R202QKR↓SVAL209 (1) |
| prM | R212SRR↓SLTV219 (0) | – |
| prM/E | P297AYS↓FNCL304 (0) | P277SMT↓MRCI284 (1) |
| E/NS1 | N784VHA↓DTGC791 (0) | M772VQA↓DSGC779 (2) |
| NS1/NS2A | R1140VNA↓YNAD1147 (0) | L1124VTA↓GHGQ1131 (1) |
| NS2A | K1327EKR↓SSAA1334 (0) | – |
| NS2A | A1334KKK↓GACL1341 (31) | – |
| NS2A/NS2B | N1367RKR↓GWPA1374 (64) | S1342KKR↓SWPL1349 (5) |
| NS2B–NS3 | Y1498TKR↓GGVL1505 (73) | K1472KQR↓AGVL1479 (11) |
| NS3pro/hel | R1686KKQ↓ITVL1693 (4) | R1659KRR↓LTIM1666 (68) |
| NS3pro/hel | K1700TRK↓ILPQ1707 (3) | K1674TKR↓YLPA1681 (31) |
| NS3pro/hel | K1716RLR↓TAVL1723 (0) | A1686IKR↓GLRT1693 (6) |
| NS3hel | A1957QRR↓GRIG1964 (2) | A1930QRR↓GRIG1937 (45) |
| NS3/NS4A | S2117GKR↓SQIG2124 (0) | A2090GRK↓SLTL2097 (1) |
| NS4A | E2243KQR↓SQTD2250 (0) | E2217KQR↓TPQD2224 (0) |
| NS4A/NS4B | A2266VAA↓NEMG2273 (0) | A2240TMA↓NEMG2247 (0) |
| NS4B/NS5 | G2522LKR↓GGAK2529 (73) | N2488TRR↓GTGN2495 (1) |
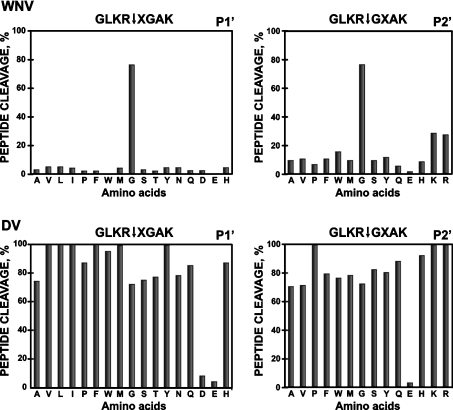
The cleavage of the peptides observed in the cleavage assay was confirmed by MS analysis (Figure 4). Thus the peptides FAM-Q102KKR↓GGTA109G-biotin (molecular mass 1689 Da) and FAM-Y1498TKR↓GGVL1505G-biotin (molecular mass 1737 Da) from the sequence of the WNV capsid C protein and from the WNV NS2B/NS3 junction region were efficiently cleaved by WNV NS2B–NS3pro and generated digest products with an expected molecular mass (FAM-QKKR, 917 Da, and FAM-YTKR, 925 Da respectively). The FAM-A1930QRR↓GRIG1937G-biotin (molecular mass 1756 Da) derived from the NS3hel sequence was also cleaved by the DV NS2B–NS3pro activity and the expected FAM-AQRR product (molecular mass 887 Da) was identified in the digest reactions. An additional confirmation of selectivity and accuracy of our approach was generated by the studies involving furin. Furin is known to be involved in the processing of prM protein of the viral polyprotein precursor [14,42–44]. In comparison with the WNV NS2B–NS3pro, furin exhibits the more restricted cleavage preferences [29,42,44]. In agreement with the cleavage preferences of furin, only the peptides HSRRSRR↓S and RSRR↓SLTV which were derived from the H209SRRSRRSLTV219 furin cleavage motif in the WNV prM protein were cleaved by furin, with 22% and 55% efficiency respectively. Similarly, furin cleaved, with a 13% efficiency, the peptide RQKR↓SVAL (amino acids 202–209) from the known furin cleavage site of the DV prM sequence. In contrast, furin was incapable of cleaving the peptides which span the NS2B/NS3 boundary (YTKRGGVL and KKQRAGVL of WNV and DV respectively), the NS3/NS4A boundary (SGKRSQIG and AGRKSLTL of WNV and DV respectively) and the NS4B/NS5 (GLKRGGAK and NARRGTGN of WNV and DV respectively) thus supporting the specificity and the accuracy of the peptide screening assay as well as our data on the role of the viral NS2B–NS3 protease in polyprotein precursor processing. The cleavage map of the polyprotein precursor of WNV and DV which is based on the peptide cleavage data is summarized in Figure 6. The sequence of the initial peptides was then modified to insert amino acid substitutions, primarily at the P3, P4 and P1′–P4′ positions of the cleavage peptides. Thus the G2522LKR↓GGAK2529 peptide from the NS4B/NS5 junction region was assayed in a positional scanning format where the P4–P1 and the P3′–P4′ positions were fixed and the P1′ and P2′ positions were each randomized with 17 and 14 amino acids (Figure 7; X represents the randomized positions). Because the library was tested at either a constant or highly similar peptide substrate concentration under the exhaustive proteolysis conditions, the relative significance of the amino acid substitutions can be directly identified. An exclusive preference for a glycine residue at both the P1′ and the P2′positions was observed with the WNV enzyme (Figure 7). In contrast, the DV NS2B–NS3pro tolerated well the presence of many types of amino acid residue, except the negatively charged aspartate and glutamate residues, at the P1′–P2′ positions.
Figure 6. The cleavage map of the WNV and the DV polyprotein precursor.
The top model shows the NS3pro cleavage sites which were predicted previously by other authors [7,10,12,13,16–19,24,26,36,38]. The lower model shows the predictions based on the present results. The peptide sequences (Table 1) which span the potential cleavage sites were synthesized and then subjected to the WNV and DV proteases. The numbers indicate the percentage efficiency of the cleavage.
Figure 7. P1′–P2′ substrate specificity of WNV and DV NS2B–NS3pro.
The P4–P1 and the P3′–P4′ positions of the GLKR↓GGAK peptide were fixed and the P1′ and P2′ positions were each randomized with 17 and 14 amino acids respectively. X represents the randomized positions. Single letter amino acid nomenclature is used.
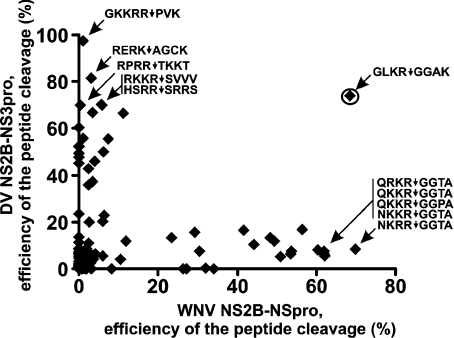
The data of a similar, albeit more extensive, analysis of 96 peptide sequences are presented in Figure 8. The purpose of this analysis, was the unbiased identification of peptide sequences which distinguish the WNV and DV proteases. Thus the (Q/N)(R/K)R↓GG(T/P)A peptides were highly selective for the WNV protease whereas the DV protease was poorly active against these peptide sequences. The REPK↓AGCK, RPRR↓TKKT, RKKR↓SVVV, HSRR↓SRRS and especially GKKRR↓PVK peptides were resistant to the WNV protease and selective for the DV NS2B–NS3pro.
Figure 8. Cleavage peptides distinguish the DV and the WNV proteases.
The efficiency of hydrolysis of 96 peptides by WNV and DV NS2B–NS3pro is shown. GLKR↓GGAK (encircled) is equally sensitive to both enzymes. The REPK↓AGCK, RPRR↓TKKT, RKKR↓SVVV, HSRR↓SRRS and especially GKKRR↓PVK peptides were selective for DV NS2B–NS3pro and the QRKR↓GGTA, QKKR↓GGTA, QKKR↓GGPA, NKKR↓GGTA and NKRR↓GGTA peptides were selective for the WNV enzyme.
DISCUSSION
The recent introduction of WNV into North America has highlighted the importance of the threat from mosquito-borne viral diseases [45]. There are currently no effective countermeasures against flaviviruses including WNV and DV. By virtue of its essential function in post-translational processing of the viral polyprotein precursor, the NS3 serine protease is a promising target for the design of flaviviral inhibitors [28,46,47]. In the present study, we report the bacterial expression, purification and substrate cleavage preferences of the homologous WNV NS2B–NS3pro in comparison with the DV type 2 NS2B–NS3pro.
A total of 300 peptides were synthesized and the efficiency of their cleavage by the WNV and DV NS2B–NS3 proteases was determined. The data generated in the present study support and extend previous observations by other laboratories [37,38,48,49]. Our results suggest that significant interactions of viral NS2B–NS3 proteases are restricted to P2–P2′. The substrate profiling study of WNV NS2B–NS3pro presented evidence that clearly supports the importance of P2–P2′ in substrate peptide cleavage as indicated by the strong dibasic preference at P1/P2 as well as the small amino acid (preferably a glycine residue) at P1′ and P2′. On the contrary, the DV enzyme could accommodate a number of amino acid residues, including the bulky hydrophobic tryptophan, phenylalanine and tyrosine residues at P1′/P2′, especially if a glycine residue is present at one of these two substrate positions. The remarkable flexibility of the DV protease permitted the design of peptide substrates which can discriminate between the closely related flaviviral enzymes. Thus the peptides GKKRRPVK, GLKRWGAK and GLKRFGAK and similar were resistant to proteolysis by the WNV enzyme but were exquisitely specific for DV NS2B–NS3pro. Conversely, the peptide NKRRGGTA was highly selective and resistant to WNV and DV NS2B–NS3pro respectively. Our recent, atomic resolution, crystallographic studies have confirmed the distinct organization of the active site cavity of the WNV two-component NS2B–NS3 protease when compared with that of the DV protease, thus corroborating our cleavage studies (A. Aleshin, S. Shiryaev, A. Strongin and R. Liddington, unpublished work).
The differences in the cleavage preferences of the flaviviral proteases are likely to have significant physiological consequences in the processing of the viral polyprotein precursor. The cleavage map of the WNV and DV precursors that was predicted, based on the peptide cleavage data, is summarized in Figure 6. The WNV protease can not cleave in trans the NS3pro/hel boundary in the precursor polyprotein with the same efficiency that the DV protease does. The pro/hel boundary, however, is readily cleaved in cis by the integral protease activity in the proteolytically potent WNV NS2B–NS3pro-hel K48A construct. In addition, in our assays the peptides that span the junction regions between NS3/NS4A (A2090GRKSLTL2097), NS4A/NS4B (A2240TMANEMG2247) and NS4B/NS5 (N2488TRRGTGN2495) of the DV polyprotein were inefficiently cleaved by the DV protease. Similarly, the NS4B/NS5 junction region peptide from the DV type 3 sequence (T2487GKRGTGS2494) was resistant to both WNV and DV proteases. Our data correlate well with the results of several previous studies. Thus relative to the peptides derived from the capsid C protein and from the NS2A/NS2B, NS2B/NS3 and NS3/NS4A boundaries the NS4B/NS5 S2488TRRGTGN2495 peptide from the DV type 2 sequence was also inefficiently cleaved by DV NS2B–NS3pro [38]. The previous data that demonstrated the partial cleavage of the biosynthetically labelled NS4B–NS5 DV precursor either by the disproportionate excess of the purified NS2B–NS3pro in vitro [18] or in the co-transfection cell-based assays [19] also indirectly support the reduced sensitivity of the NS4B/NS5 boundary sequence to proteolysis by NS2B–NS3. There is an additional possible explanation for the differences of our results obtained with short peptide substrates relative to the previous data of others who used long polyprotein precursor proteins. Because the biosynthetically labelled NS4B–NS5 substrates were prepared as the membrane-associated proteins these earlier data also support the role of membranes in specific presentation of the cleavage site to the protease [18,19,50].
The precise physiological consequences of our biochemical, in vitro, findings are unclear. The differential rate of cleavage at the major cleavage sites may be intertwined with a required, ordered processing of the flaviviral polyprotein precursor, yielding sufficient intermediates with the desired function for viral assembly. Overall, our data indicate that even the suboptimal sequence motifs, because of their specific presentation into the active site of the protease, may be cleaved in cis in the course of the polyprotein precursor processing in vivo.
Evidence suggests that the NS3 protease is essential for the cleavage of the flaviviral polyprotein at least at the NS2A/NS2B, NS2B/NS3, NS3/NS4A, NS4A/NS4B and NS4B/NS5 boundaries and also within the sequence of capsid C protein (Table 3). Because of its furin-like, restricted, substrate cleavage specificity [18,29,37,44], the NS3pro requires the presence of a positively charged arginine residue and either an arginine or lysine residue at the P1 and P2 positions respectively, for the efficient cleavage of scissile bonds. These requirements result in the conservation of the natural cleavage sites in the flaviviral polyprotein precursor. The only exception is the cleavage site at the NS2B–NS3 boundary, in which a glutamine residue occupies the P2 position in all four DV serotypes. In the polyprotein precursor, the dibasic (Lys/Arg)–Arg motif is followed by a small or polar residue including predominantly glycine and serine. In the West Nile polyprotein, a glycine residue most frequently occupies both the P1′ and the P2′ positions. A similar pattern is also characteristic for JEV (Japanese encephalitis virus). In turn, in YFV (yellow fever virus) and in all four DV serotypes, a serine residue and in several instances an alanine and threonine residue are at the P1′ position. When a serine, alanine or threonine residue occupies P1′, multiple amino acid types (serine, threonine, alanine, valine, leucine, isoleucine and tryptophan) are allowed at the P2′ position in DV. As long as a glycine residue occupies the P1′ at the NS4B–NS5 boundary and the P2′ at the NS2B–NS3 boundary in DV, the other P′ position may be occupied by a serine, alanine or threonine residue. This specificity pattern correlates well with the results of the peptide cleavage screens and indicate that the WNV NS3pro specifically developed the capability for cleaving the Gly–Gly motifs while the DV enzyme adopted a less restricted specificity to process the natural cleavage sites in the polyprotein precursor.
Table 3. The sequence of the natural cleavage sites of the NS3 protease in the capsid protein C and at the NS2A/NS2B, NS2B/NS3, NS3/NS4A, NS4A/NS4B and NS4B/NS5 boundaries of the polyprotein precursor.
WNV, West Nile virus (GenBank™ accession number P06935); JEV, Japanese encephalitis virus (GenBank™ accession number P19110); YFV, Yellow fever virus (GenBank™ accession number P19901); DV1-4, Dengue virus serotypes 1-4 (GenBank™ accession number P33478, P29990, P27915 and P09866 respectively). Ser102 (capsid protein C) and Asn2488 (the NS4B/NS5 boundary) are frequently replaced by Thr102 and Ser2488 respectively, in certain subtypes of DV2 (both residues are underlined below).
| Capsid C | NS2A/NS2B | NS2B–NS3 | NS3/NS4A | NS4B/NS5 | |
|---|---|---|---|---|---|
| WNV | Q102KKR↓GGTA109 | N1367RKR↓GWPA1374 | Y1498TKR↓GGVL1505 | S2117GKR↓SQIG2124 | G2522LKR↓GGAK2529 |
| JEV | Q102NKR↓GGNE109 | N1370KKR↓GWPA1377 | T1501TKR↓GGVF1508 | A2120GKR↓SAVS2127 | S2524LKR↓GRPG2531 |
| YFV | R98 KRR↓SHDV105 | F1351GRR↓SIPV1358 | G1481ARR↓SGDV1488 | E2104GRR↓GAAE2111 | T2503GRR↓GSAN2510 |
| DV1 | R97 RKR↓SVTM104 | W1341GRK↓SWPL1348 | K1471KQR↓SGVL1478 | A2090GRR↓SVSG2097 | G2489GRR↓GTGA2496 |
| DV2 | R97 RRR↓SAGV104 | S1342KKR↓SWPL1349 | K1472KQR↓AGVL1479 | A2090GRK↓SLTL2097 | N2488TRR↓GTGN2495 |
| DV3 | K97 RKK↓TSLC104 | L1340KRR↓SWPL1347 | Q1470TQR↓SGVL1477 | A2089GRK↓SIAL2096 | T2487GKR↓GTGS2494 |
| DV4 | G96 RKR↓STIT103 | A1341SRR↓SWPL1348 | K1471TQR↓SGAL1478 | S2089GRK↓SITL2096 | T2484PRR↓GTGT2491 |
We believe that the present studies have produced a set of valuable research tools for uncovering the structural and functional mechanisms which modulate the viral proteinase activity and the cleavage specificity and that our data represent a valuable resource for the design of highly selective, fluorescence quenched, peptide derivatives for studies of flaviviral proteinases. We now have a clear starting point for developing novel diagnostic assays and therapeutics aimed at a broad range of flaviviruses. In addition, the unique cleavage preferences of the WNV and DV proteases promise novelty for the design of potent inhibitors that may be selective for the flaviviral proteases over proteases of the host cell and thus provide a solid base for follow-on structural and functional studies.
Acknowledgments
This work was supported by NIH (National Institutes of Health) Grants CA83017 and CA7747 (to A.Y.S.), AI056869 (to M.L.) and RR020843 (J.W.S. and A.Y.S.). We thank Veronica Shevchenko and John Hachmann for their professional assistance with the peptide synthesis and the follow-on experiments, and Peter Melnyk, Chanfeng Zhao, Sergey Bibikov and Anu Srinivasan for their suggestions and discussion.
References
- 1.Chambers T. J., Diamond M. S. Pathogenesis of flavivirus encephalitis. Adv. Virus Res. 2003;60:273–342. doi: 10.1016/S0065-3527(03)60008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samuel C. E. Host genetic variability and West Nile virus susceptibility. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11555–11557. doi: 10.1073/pnas.202448899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukhopadhyay S., Kuhn R. J., Rossmann M. G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 4.Chambers T. J., Droll D. A., Tang Y., Liang Y., Ganesh V. K., Murthy K. H., Nickells M. Yellow fever virus NS2B–NS3 protease: characterization of charged-to-alanine mutant and revertant viruses and analysis of polyprotein-cleavage activities. J. Gen. Virol. 2005;86:1403–1413. doi: 10.1099/vir.0.80427-0. [DOI] [PubMed] [Google Scholar]
- 5.Chambers T. J., Nestorowicz A., Amberg S. M., Rice C. M. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B–NS3 complex formation, and viral replication. J. Virol. 1993;67:6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chappell K. J., Nall T. A., Stoermer M. J., Fang N. X., Tyndall J. D., Fairlie D. P., Young P. R. Site-directed mutagenesis and kinetic studies of the West Nile Virus NS3 protease identify key enzyme–substrate interactions. J. Biol. Chem. 2005;280:2896–2903. doi: 10.1074/jbc.M409931200. [DOI] [PubMed] [Google Scholar]
- 7.Clum S., Ebner K. E., Padmanabhan R. Co-translational membrane insertion of the serine proteinase precursor NS2B–NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J. Biol. Chem. 1997;272:30715–30723. doi: 10.1074/jbc.272.49.30715. [DOI] [PubMed] [Google Scholar]
- 8.Falgout B., Miller R. H., Lai C. J. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B–NS3 protease activity. J. Virol. 1993;67:2034–2042. doi: 10.1128/jvi.67.4.2034-2042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falgout B., Pethel M., Zhang Y. M., Lai C. J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of Dengue virus nonstructural proteins. J. Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keelapang P., Sriburi R., Supasa S., Panyadee N., Songjaeng A., Jairungsri A., Puttikhunt C., Kasinrerk W., Malasit P., Sittisombut N. Alterations of pr-M cleavage and virus export in pr-M junction chimeric Dengue viruses. J. Virol. 2004;78:2367–2381. doi: 10.1128/JVI.78.5.2367-2381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin C., Pragai B. M., Grakoui A., Xu J., Rice C. M. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J. Virol. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Preugschat F., Yao C. W., Strauss J. H. In vitro processing of Dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J. Virol. 1990;64:4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shafee N., AbuBakar S. Dengue virus type 2 NS3 protease and NS2B–NS3 protease precursor induce apoptosis. J. Gen. Virol. 2003;84:2191–2195. doi: 10.1099/vir.0.19022-0. [DOI] [PubMed] [Google Scholar]
- 14.Stadler K., Allison S. L., Schalich J., Heinz F. X. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 1997;71:8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stocks C. E., Lobigs M. Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B–3 protease for efficient processing requires determinants in C, the signal peptide, and prM. J. Virol. 1998;72:2141–2149. doi: 10.1128/jvi.72.3.2141-2149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamshchikov V. F., Compans R. W. Processing of the intracellular form of the West Nile virus capsid protein by the viral NS2B–NS3 protease: an in vitro study. J. Virol. 1994;68:5765–5771. doi: 10.1128/jvi.68.9.5765-5771.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamshchikov V. F., Trent D. W., Compans R. W. Upregulation of signalase processing and induction of prM-E secretion by the flavivirus NS2B–NS3 protease: roles of protease components. J. Virol. 1997;71:4364–4371. doi: 10.1128/jvi.71.6.4364-4371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yusof R., Clum S., Wetzel M., Murthy H. M., Padmanabhan R. Purified NS2B/NS3 serine protease of Dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J. Biol. Chem. 2000;275:9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 19.Cahour A., Falgout B., Lai C. J. Cleavage of the Dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B–NS3, whereas NS4A/NS4B may be processed by a cellular protease. J. Virol. 1992;66:1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yon C., Teramoto T., Mueller N., Phelan J., Ganesh V. K., Murthy K. H., Padmanabhan R. Modulation of the nucleoside triphosphatase/RNA helicase and 5′-RNA triphosphatase activities of Dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J. Biol. Chem. 2005;280:27412–27419. doi: 10.1074/jbc.M501393200. [DOI] [PubMed] [Google Scholar]
- 21.Kang L. W., Cho H. S., Cha S. S., Chung K. M., Back S. H., Jang S. K., Oh B. H. Crystallization and preliminary X-ray crystallographic analysis of the helicase domain of hepatitis C virus NS3 protein. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:121–123. doi: 10.1107/s0907444997008883. [DOI] [PubMed] [Google Scholar]
- 22.Mackintosh S. G., Lu J. Z., Jordan J. B., Harrison M. K., Sikora B., Sharma S. D., Cameron C. E., Raney K. D., Sakon J. Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus. J. Biol. Chem. 2006;281:3528–3535. doi: 10.1074/jbc.M512100200. [DOI] [PubMed] [Google Scholar]
- 23.Tai C. L., Pan W. C., Liaw S. H., Yang U. C., Hwang L. H., Chen D. S. Structure-based mutational analysis of the hepatitis C virus NS3 helicase. J. Virol. 2001;75:8289–8297. doi: 10.1128/JVI.75.17.8289-8297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nall T. A., Chappell K. J., Stoermer M. J., Fang N. X., Tyndall J. D., Young P. R., Fairlie D. P. Enzymatic characterization and homology model of a catalytically active recombinant West Nile virus NS3 protease. J. Biol. Chem. 2004;279:48535–48542. doi: 10.1074/jbc.M406810200. [DOI] [PubMed] [Google Scholar]
- 25.Brinkworth R. I., Fairlie D. P., Leung D., Young P. R. Homology model of the dengue 2 virus NS3 protease: putative interactions with both substrate and NS2B cofactor. J. Gen. Virol. 1999;80:1167–1177. doi: 10.1099/0022-1317-80-5-1167. [DOI] [PubMed] [Google Scholar]
- 26.Leung D., Schroder K., White H., Fang N. X., Stoermer M. J., Abbenante G., Martin J. L., Young P. R., Fairlie D. P. Activity of recombinant Dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J. Biol. Chem. 2001;276:45762–45771. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- 27.Niyomrattanakit P., Winoyanuwattikun P., Chanprapaph S., Angsuthanasombat C., Panyim S., Katzenmeier G. Identification of residues in the Dengue virus type 2 NS2B cofactor that are critical for NS3 protease activation. J. Virol. 2004;78:13708–13716. doi: 10.1128/JVI.78.24.13708-13716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beasley D. W. Recent advances in the molecular biology of West Nile virus. Curr. Mol. Med. 2005;5:835–850. doi: 10.2174/156652405774962272. [DOI] [PubMed] [Google Scholar]
- 29.Shiryaev S. A., Ratnikov B. I., Chekanov A. V., Sikora S., Rozanov D. V., Godzik A., Wang J., Smith J. W., Huang Z., Lindberg I., et al. Cleavage targets and the D-arginine-based inhibitors of the West Nile virus NS3 processing proteinase. Biochem. J. 2006;393:503–511. doi: 10.1042/BJ20051374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kacprzak M. M., Peinado J. R., Than M. E., Appel J., Henrich S., Lipkind G., Houghten R. A., Bode W., Lindberg I. Inhibition of furin by polyarginine-containing peptides: nanomolar inhibition by nona-D-arginine. J. Biol. Chem. 2004;279:36788–36794. doi: 10.1074/jbc.M400484200. [DOI] [PubMed] [Google Scholar]
- 31.Lebl M. New technique for high-throughput synthesis. Bioorg. Med. Chem. Lett. 1999;9:1305–1310. doi: 10.1016/s0960-894x(99)00176-6. [DOI] [PubMed] [Google Scholar]
- 32.Lebl M., Hachmann J. High-throughput peptide synthesis. Methods Mol. Biol. 2005;298:167–194. doi: 10.1385/1-59259-877-3:167. [DOI] [PubMed] [Google Scholar]
- 33.Hachmann J., Lebl M. Alternative to piperidine in Fmoc solid-phase synthesis. J. Comb. Chem. 2006;8:149. doi: 10.1021/cc050123l. [DOI] [PubMed] [Google Scholar]
- 34.Fischer R., Mader O., Jung G., Brock R. Extending the applicability of carboxyfluorescein in solid-phase synthesis. Bioconjugate Chem. 2003;14:653–660. doi: 10.1021/bc025658b. [DOI] [PubMed] [Google Scholar]
- 35.King D. S., Fields C. G., Fields G. B. A cleavage method which minimizes side reactions following Fmoc solid phase peptide synthesis. Int. J. Pept. Protein Res. 1990;36:255–266. doi: 10.1111/j.1399-3011.1990.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 36.Wu C. F., Wang S. H., Sun C. M., Hu S. T., Syu W. J. Activation of dengue protease autocleavage at the NS2B–NS3 junction by recombinant NS3 and GST-NS2B fusion proteins. J. Virol. Methods. 2003;114:45–54. doi: 10.1016/j.jviromet.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Li J., Lim S. P., Beer D., Patel V., Wen D., Tumanut C., Tully D. C., Williams J. A., Jiricek J., Priestle J. P., et al. Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetrapeptide and octapeptide substrate libraries. J. Biol. Chem. 2005;280:28766–28774. doi: 10.1074/jbc.M500588200. [DOI] [PubMed] [Google Scholar]
- 38.Niyomrattanakit P., Yahorava S., Mutule I., Mutulis F., Petrovska R., Prusis P., Katzenmeier G., Wikberg J. E. Probing the substrate specificity of the Dengue virus type 2 NS3 serine protease by using internally quenched fluorescent peptides. Biochem. J. 2006;397:203–211. doi: 10.1042/BJ20051767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hachmann J., Lebl M. Search for optimal coupling reagent in multiple peptide synthesizer. Biopolymers. 2006;84:340–347. doi: 10.1002/bip.20482. [DOI] [PubMed] [Google Scholar]
- 40.Lebl M., Pokorny V., Krchnak V. Simple tools for resin distribution. J. Comb. Chem. 2005;7:42–45. doi: 10.1021/cc049930v. [DOI] [PubMed] [Google Scholar]
- 41.Kozlov I. A., Melnyk P. C., Zhao C., Hachmann J. P., Shevchenko V., Srinivasan A., Barker D. L., Lebl M. A method for rapid protease substrate evaluation and optimization. Comb. Chem. High Throughput Screen. 2006;9:481–487. doi: 10.2174/138620706777698535. [DOI] [PubMed] [Google Scholar]
- 42.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell. Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elshuber S., Allison S. L., Heinz F. X., Mandl C. W. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 2003;84:183–191. doi: 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- 44.Seidah N. G. Unexpected similarity between the cytosolic West Nile virus NS3 and the secretory furin-like serine proteinases. Biochem. J. 2006;393:e1–e3. doi: 10.1042/BJ20051787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayes C. G. West Nile virus: Uganda, 1937, to New York City, 1999. Ann. NY Acad. Sci. 2001;951:25–37. doi: 10.1111/j.1749-6632.2001.tb02682.x. [DOI] [PubMed] [Google Scholar]
- 46.Erbel P., Schiering N., D'Arcy A., Renatus M., Kroemer M., Lim S. P., Yin Z., Keller T. H., Vasudevan S. G., Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 47.Melino S., Fucito S., Campagna A., Wrubl F., Gamarnik A., Cicero D. O., Paci M. The active essential CFNS3d protein complex. FEBS J. 2006;273:3650–3662. doi: 10.1111/j.1742-4658.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 48.Khumthong R., Angsuthanasombat C., Panyim S., Katzenmeier G. In vitro determination of dengue virus type 2 NS2B–NS3 protease activity with fluorescent peptide substrates. J. Biochem. Mol. Biol. 2002;35:206–212. doi: 10.5483/bmbrep.2002.35.2.206. [DOI] [PubMed] [Google Scholar]
- 49.Khumthong R., Niyomrattanakit P., Chanprapaph S., Angsuthanasombat C., Panyim S., Katzenmeier G. Steady-state cleavage kinetics for dengue virus type 2 NS2B–NS3(pro) serine protease with synthetic peptides. Protein Pept. Lett. 2003;10:19–26. doi: 10.2174/0929866033408228. [DOI] [PubMed] [Google Scholar]
- 50.Lin C., Chambers T. J., Rice C. M. Mutagenesis of conserved residues at the yellow fever virus 3/4A and 4B/5 dibasic cleavage sites: effects on cleavage efficiency and polyprotein processing. Virology. 1993;192:596–604. doi: 10.1006/viro.1993.1076. [DOI] [PubMed] [Google Scholar]