Abstract
Toxofilin is a 27 kDa protein isolated from the human protozoan parasite Toxoplasma gondii, which causes toxoplasmosis. Toxofilin binds to G-actin, and in vitro studies have shown that it controls elongation of actin filaments by sequestering actin monomers. Toxofilin affinity for G-actin is controlled by the phosphorylation status of its Ser53, which depends on the activities of a casein kinase II and a type 2C serine/threonine phosphatase (PP2C). To get insights into the functional properties of toxofilin, we undertook a structure–function analysis of the protein using a combination of biochemical techniques. We identified a domain that was sufficient to sequester G-actin and that contains three peptide sequences selectively binding to G-actin. Two of these sequences are similar to sequences present in several G- and F-actin-binding proteins, while the third appears to be specific to toxofilin. Additionally, we identified two toxofilin domains that interact with PP2C, one of which contains the Ser53 substrate. In addition to characterizing the interacting domains of toxofilin with its partners, the present study also provides information on an in vivo-based approach to selectively and competitively disrupt the protein–protein interactions that are important to parasite motility.
Keywords: actin, peptide-spot mapping assay, protein–protein interaction, pyrene actin assay, serine/threonine phosphatase, toxofilin
Abbreviations: PP2C, type 2C serine/threonine phosphatase; GST, glutathione S-transferase; IPTG, isopropyl β-D-thiogalactoside; HPTR, histidine patch thioredoxine; IAEDANS, N-iodoacetyl-N′-(5-sulfo-1-naphthyl)ethylenediamine; TBS, Tris buffered saline; HRP, horseradish peroxidase
INTRODUCTION
The human protozoan parasite Toxoplasma gondii, together with other members of the Apicomplexa phylum, invade their host cell by an active process that markedly differs from the passive uptake induced by invasive bacteria [1]. Following host cell entry, the parasite resides within a sub-cellular compartment called the parasitophorous vacuole. The parasite-driven remodelling of this compartment allows molecular exchanges to take place between the host cell and the parasite, thereby promoting subsequent parasite development [2,3]. The massive multiplication of T. gondii tachyzoites within this vacuole ends by host cell lysis and active parasite egress, which leads to parasite dissemination [4,5]. These events are associated with potentially severe or lethal pathologies, especially when they occur in the brain of immunocompromised humans or during fetal development (for a review see [6,7]).
The tachyzoite actively enters its host cell by developing a force that relies on actin polymerization [8] and myosin activation [9,10]. This force is generated upon contact with the host cell, but the signalling underlying parasite actin polymerization remains unclear. A key role has been ascribed to a micronemal protein, MIC2, which is secreted on the parasite surface upon parasite–host cell contact, links the parasite motor to the extracellular cues and is translocated rearwards by the motor as the parasite moves forward [11,12]. The glycolytic enzyme aldolase has been found to selectively associate with the MIC2 tail and with actin, thereby forming a direct bridge between MIC2 and actin [13].
We previously identified a 27 kDa T. gondii protein called toxofilin, which can bind to both mammalian and parasite actin. In vitro assays performed with mammalian actin demonstrated that toxofilin sequesters actin monomers and thus controls filament elongation [14]. We also characterized a type 2C serine/threonine phosphatase (PP2C) that co-purifies with the actin–toxofilin complex and selectively dephosphorylates toxofilin at Ser53, thereby increasing toxofilin affinity to G-actin [15].
A central question remains as to whether toxofilin acts on mammalian actin, Toxoplasma actin or both. Several results argue in favour of toxofilin targeting host cell actin. We have described a major apical localization of toxofilin in tachyzoites, in particular in intracellular stages of the parasite, an observation consistent with the recent identification of toxofilin as a rhoptry protein following a proteomic analysis of rhoptry contents [16]. Indeed, toxofilin, which carries an N-terminal peptide signal sequence, is likely to be secreted during cell invasion and to act locally on the host cell cortical actin during parasite penetration of the cell. On the other hand, we cannot also exclude a role for toxofilin on parasite actin dynamics. Our immunolocalization studies showed that toxofilin is also located beneath the plasma membrane and at the posterior end of invading tachyzoites. Recent data obtained with tagged versions of toxofilin expressed via its own regulatory sequences confirmed these observations (results not shown and [16]).
In the present study, using a combination of biochemical techniques, we identified the interacting domains of toxofilin with both partners, i.e. PP2C and mammalian actin. Our findings show that besides controlling actin polymerization by sequestering actin monomers via its first coil–coiled region, toxofilin also binds to G-actin through its N-terminal domain.
EXPERIMENTAL
Preparation of recombinant proteins
We expressed recombinant full and truncated toxofilin in the BL21 strain of Escherichia coli using the GST (glutathione S-transferase) Gene Fusion System (GE Healthcare U.K.). The full-length toxofilin was cloned into pGex6-p3 as described in [14] while CC1, CC1A, CC1Aa, CC1Ab, CC1B, CC1BCC2, NoCC and CC2 domains of toxofilin were prepared as follows (Figure 1A). The CC1 nucleotidic sequence was amplified using the forward primer 5′-GGCCGGATCCCAACAGGAACTAGGGCTGCTC-3′ containing a BamHI site (underlined) and the reverse primer 5′-GGCCGTCGACCCTCTGCTCGTTGAGGATTTG-3′ containing a SalI site (underlined). For CC1A, the forward primer was the same as for CC1 and the reverse primer was 5′-GGCCGTCGACCTCAGTTGCGAAAGATCCCTC-3′ (the SalI site is underlined). For CC1B, the forward primer was 5′-GGCCGGATCCCCGGAAACGAAGGCTTTGC-3′ (the BamHI site is underlined) and the reverse primer was the same as for CC1. For CC1Aa, the forward primer was the same as for CC1 and the reverse primer was 5′-GGCCGTCGACCGTGGCTCTGAGAATTTCGTC-3′ (the SalI site is underlined). For CC1Ab, the forward primer was 5′-GGCCGGATCCCAAAATTTGGACCTCAGGAAGTAC-3′ (the BamHI site is underlined) and the reverse primer was the same as for CC1A. For CC1BCC2, the forward primer was the same as for CC1B and the reverse primer was the same as for toxofilin. For NoCC, the forward primer was the same as for toxofilin and the reverse primer was 5′-GGCCGTCGACGCGTGCTGCGACGGAGGG-3′(the SalI site is underlined). Finally, CC2 was amplified using the forward primer 5′-GGCCGGATCCGATGCGAGTGGAGCATTAC-3′ (the BamHI site is underlined) and the same reverse primer as for toxofilin. The PCR products were then ligated into the BamHI and SalI sites of a pGex6-p3 polylinker. Protein expression was induced with either 0.2 mM (CC1, CC1A, CC1Aa, CC1Ab, CC1B and CC1BCC2) or 0.1 mM (NoCC, CC2 and toxofilin) IPTG (isopropyl β-D-thiogalactoside). Recombinant proteins were purified from bacteria lysed in PBS supplemented with 0.5% (v/v) Triton X-100 with or without 0.5% (v/v) SB3-14 (myristyl sulfobetaine). After centrifugation at 20000 g for 20 min at 4 °C, supernatants were incubated with Glutathione–Sepharose (GE Healthcare) for 3 h at 4 °C. The recombinant tagged-polypeptides were eluted in 50 mM Tris/HCl (pH 8.0), 50 mM NaCl and 10 mM reduced glutathione while in some cases, tags were cleaved off using PreScission Protease (GE Healthcare) for 4 h at 4 °C. The HPTR (histidine patch thioredoxine)–PP2C (rPP2C) was prepared as described in [15] using Ni-NTA column chromatography (Qiagen). Following purification, polypeptides were dialysed against the buffer used for subsequent binding, titrated, supplemented with protease inhibitors and kept on ice until use.
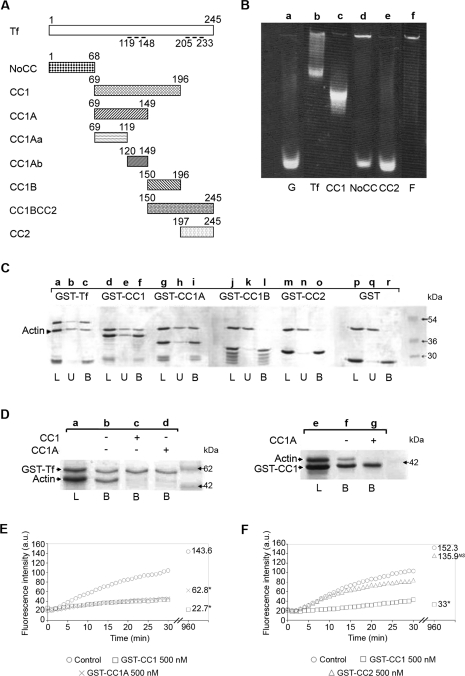
Figure 1. Toxofilin binds to G-actin via the CC1A domain.
(A) Schematic representation of full-length toxofilin and toxofilin domains used throughout the present study. Amino acids are numbered on top of the schemes. Dashed lines represent the two coil–coiled domains. Tf, toxofilin. (B) A 7.5% acrylamide native gel assay was performed using 50 μg of IAEDANS-labelled G-actin incubated without (lane a) or with equimolar amounts of recombinant GST–toxofilin (lane b), GST–CC1 (lane c), GST–NoCC (lane d) or GST–CC2 (lane e). F-actin was loaded as a control (lane f). (C) G-actin binding assays were performed on immobilized GST–toxofilin (GST–Tf; lanes a–c), GST–CC1 (lanes d–f), GST–CC1A (lanes g–i), GST–CC1B (lanes j–l), GST–CC2 (lanes m–o) and GST alone (lanes p–r). Following overnight incubation, the resin was washed and bound proteins were eluted in SDS sample buffer prior to SDS/PAGE and Coomassie Blue staining. L indicates the total load, U indicates the total unbound fraction and B indicates the total bound fraction. The arrowhead indicates the position of G-actin. (D) Actin-binding competition assays were carried out on immobilized GST–toxofilin (GST–Tf) using G-actin preincubated without (lane b) or with CC1 (lane c) or CC1A (lane d) and on immobilized GST–CC1 using G-actin preincubated or not with CC1A (lanes e–g). Samples were treated as described for (C). L indicates the total load and B indicates the total bound fraction. The presence of the competitive fragment is indicated above the gel photograph. (E) Pyrene actin polymerization kinetics in the presence or absence of 500 nM GST–CC1 (□) and GST–CC1A (×) polypeptides using pyrene actin. Numbers on the right of the curves represent the actin steady-state values obtained 16 h after the assay. *The difference with the control values (○) are statistically significant at P=0.05 (Student's t test). a.u., arbitrary units. (F) Pyrene actin polymerization kinetics in the presence or absence of 500 nM GST–CC1 (□) and GST–CC2 (△) polypeptides using pyrene actin. Numbers on the right of the curves represent the actin steady-state values obtained 16 h after the assay. *The differences with the control values (○) are statistically significant at P=0.05 (Student's t test). NS, not significant; a.u., arbitrary units.
Native gel assays
Native gel electrophoresis was carried out as described in [14] with a 7.5% acrylamide gel using G-actin prepared from F-actin labelled with 500 μM IAEDANS [N-iodoacetyl-N′-(5-sulfo-1-naphthyl)ethylenediamine; Molecular Probes]. IAEDANS-labelled G-actin (50 μg) was incubated with or without equimolar amounts of recombinant GST–toxofilin, GST–CC1, GST–NoCC or GST–CC2 for 1 h at room temperature prior to electrophoresis. F-actin was used as a control.
G-actin binding assays
Rabbit muscle actin was purified from muscle acetone powder (Sigma) and prepared as described in [17]. It was diluted to 55 μM in buffer A [2 mM Tris/HCl (pH 8.0), 0.2 mM Na2ATP, 0.1 mM CaCl2, 0.5 mM 2-mercaptoethanol and 0.005% NaN3]. Purified rabbit muscle G-actin [4.2 μg (2 μM)] was incubated overnight at 4 °C with 4 μM of GST fusion proteins prepared in 50 μl of buffer B [10 mM Tris/HCl (pH 8.0), 150 mM NaCl and 0.1 mM CaCl2]. A 50% (v/v) slurry of Glutathione–Sepharose (30 μl) in buffer B was added to the mix which was supplemented with 1 mM dithiothreitol and 0.05% (v/v) Tween 20 (final concentrations) and incubated for 3 h at 4 °C. The unbound fraction was collected by centrifugation and the beads were washed with buffer B supplemented with 0.05% (v/v) Tween 20. SDS eluate and fraction samples were analysed by SDS/PAGE [18] and Coomassie Blue staining. Western blotting using the anti-C4 actin monoclonal antibodies (Chemicon International) was performed as described in [14] except that the C4 antibodies were used at a 1:1000 dilution.
Competition assays for binding to G-actin
Purified rabbit muscle actin [4.2 μg (2 μM)] was incubated with or without 14.5 μg (20 μM) of clarified CC1, 9 μg (20 μM) of CC1A, 5.6 μg (20 μM) of CC1Aa, 3.3 μg (20 μM) of CC1Ab or a mix of CC1Aa and CC1Ab in 50 μl of buffer B for 1 h at 4 °C. CC1–actin and CC1A–actin mixes were added to 10.8 μg (4 μM) GST–toxofilin while CC1A–actin, CC1Aa–actin and CC1Ab–actin mixes were also added to 8.2 μg (4 μM) GST–CC1. All reactions were incubated overnight at 4 °C. Samples were then treated as described for the G-actin binding assays. Eluate and unbound fractions were analysed by SDS/PAGE and Coomassie Blue staining. Controls were performed with CC1 and CC1A incubated with Glutathione–Sepharose under the experimental conditions.
Actin pyrene assays and polymerization assays
Three separate pyrene assays were performed as described in [14] using 2 μM of 20% labelled G-actin. For additional polymerization assays, 12.6 μg (10 μM) of purified rabbit muscle G-actin was incubated with 12.3 μg (10 μM) of clarified GST–CC1, 10.3 μg (10 μM) of GST–NoCC, 10.8 μg (10 μM) of GST–CC1A, 9.8 μg (10 μM) GST–CC1Aa or 9.1 μg (10 μM) of GST–CC1Ab or the toxofilin domains without their GST tags in 30 μl of buffer A. Then, 100 mM KCl, 2 mM MgCl2 and 1 mM Na2ATP were added to induce polymerization of G-actin overnight at 4 °C. Unpolymerized and polymerized actins were separated following ultracentrifugation at 70000 rev./min for 30 min at 4 °C (rotor ILA 120.2, Beckman Coulter) and both fractions were analysed by SDS/PAGE and Coomassie Blue staining. In some experiments, 12.6 μg (10 μM) of G-actin was incubated with a mix of 4.4 μg (10 μM) of CC1 and 14.7 μg (10 μM) of rPP2C prior to inducing actin polymerization. To examine the potential effect of NoCC on actin in G- conditions, similar assays were conducted without adding the polymerization conditions.
PP2C-binding assays
HPTR-tagged PP2C [3.9 μg (2 μM)] was immobilized on 10 μl of a 50% (v/v) slurry of Ni-NTA–agarose (Qiagen) washed in buffer C [20 mM Tris/HCl (pH 7.5), 150 mM NaCl, 1 mM MgCl2 and 0.01% (v/v) Triton X-100] for 2 h at 4 °C. Bound rPP2C was then incubated with the different domains of toxofilin for 1 h at 4 °C and the putative complexes were isolated by centrifugation (400 g, 4 min at 4 °C). The unbound fractions were recovered and the beads were washed with a 10 times volume of buffer D [20 mM Tris/HCl (pH 7.5), 200 mM NaCl, 1 mM MgCl2 and 0.01% (v/v) Triton X-100]. SDS sample buffer eluates and fractions were then analysed by SDS/PAGE and Coomassie Blue staining.
PP2C–CC1–actin complex binding assays
rPP2C bound to Ni-NTA–agarose [3.9 μg (2 μM)] was incubated either first with the preformed CC1–actin complex in buffer A (1 h at 4 °C) or sequentially with 2.3 μg (4 μM) of CC1 in buffer B (1 h at 4 °C) followed by 3.4 μg (2 μM) of clarified G-actin in buffer B (1 h at 4 °C). Following incubation, the resin was washed in buffer D and treated as described for the PP2C-binding assay.
Peptide-spot mapping
In order to identify the important amino acid residues of the CC1A polypeptide that could account for G-actin binding, we performed a peptide-spot assay using synthetic overlapping dodecapeptides spotted onto a cellulose membrane (Intavis AG). The entire sequence of CC1A was represented, and each peptide differed from the following by a shift of two amino acids. Before use, the membranes were wetted in distilled water then in TBS-T [Tris-buffered saline supplemented with 0.1% (v/v) Tween 20] and saturated in TBS-T containing 3% (w/v) non-fat dried skimmed milk and 3% (w/v) BSA for 2 h at room temperature (23 °C). To detect toxofilin/CC1A–G-actin interacting sequences, membranes were then incubated with 1 μg/ml of biotinylated G-actin in buffer A supplemented with 1% (w/v) non-fat dried skimmed milk overnight at 4 °C. After several washes in TBS-T, the membranes were incubated with streptavidin–HRP (horseradish peroxidase) at a 1:2000 dilution (Upstate) for 2 h at room temperature. To detect toxofilin–PP2C interacting sequences, the toxofilin membranes were incubated overnight at 4 °C with 4 μg/ml of rPP2C in TBS-T supplemented with 1% (w/v) non-fat dried skimmed milk. Membranes were washed in TBS-T, incubated first with an affinity purified polyclonal anti-T. gondii PP2C antibody at a 1:4000 dilution for 2 h at room temperature and secondly with anti rabbit-HRP secondary antibody (Jackson ImmunoResearch) at a 1:10000 dilution for 2 h at room temperature. Following extensive washes in TBS-T and TBS-T containing 0.2% (v/v) Tween 20, positive spots were detected using the SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnologies).
RESULTS
Toxofilin binds to G-actin via the CC1A domain and sequesters G-actin
To study the structure–function relationships of toxofilin, we performed a series of assays with purified mammalian actin. Sequence analysis of toxofilin predicts that the protein is globular with two coil–coiled regions (contained in the CC1 and CC2 domains respectively, see Figure 1A) and that it carries a signal peptide sequence at the N-terminal end. To map the toxofilin domains involved in actin binding, we first produced several polypeptides encompassing or not the coil–coiled regions (Figure 1A) and tested their binding properties to G-actin using pull-down assays and native gel electrophoresis. Since toxofilin was originally isolated using a native gel assay with IAEDANS-labelled actin [14], we used the same assay conditions to define the toxofilin regions that bind to actin. When fluorescently labelled G-actin (Figure 1B, lane a) was incubated with either full-length recombinant toxofilin or toxofilin truncated polypeptides CC1 (14.6 kDa), CC2 (5.5 kDa) or NoCC (7.5 kDa) prior to native gel electrophoresis, only toxofilin and CC1 induced a clear shift in actin migration (Figure 1B, lanes b and c). In contrast, NoCC and CC2 did not similarly alter actin migration (Figure 1B, lanes d and e). Interestingly, NoCC addition to G-actin prevented part of the G-actin pool from entering the 7.5% acrylamide gel in a similar manner to when F-actin was loaded onto the gel (Figure 1B, lane f). This suggested that NoCC could also bind to G-actin and promote actin oligomerization or nucleation. Together these results indicated that the toxofilin binding site(s) for G-actin is located in the CC1 polypeptide and is sufficient to capture actin.
To narrow down the putative minimal G-actin-binding domain, the 14.5 kDa CC1 domain was cut into 8.9 kDa CC1A and 5.5 kDa CC1B polypeptides and we performed chromatography on Glutathione–Sepharose of GST fusion polypeptides pre-incubated with G-actin. As illustrated in Figure 1(C), only GST–toxofilin (lanes a–c), GST–CC1 (lanes d–f) and GST–CC1A (lanes g–i), but not GST–CC1B, GST–CC2 or GST (lanes j–r), were able to capture G-actin.
To confirm that the 8.9 kDa CC1A polypeptide could bind G-actin, we tested whether it could compete with either full-length toxofilin or CC1 for binding to G-actin. Figure 1(D) shows a complete inhibition of actin binding to GST–toxofilin in the presence of a 5-fold molar excess of CC1 or CC1A (lanes b–d). Additionally, actin binding to GST–CC1 was fully prevented by a 5-fold molar excess of CC1A (lanes f and g). As controls, actin, CC1 and CC1A did not bind to the gluthatione matrix under our conditions (results not shown). Together, these data demonstrate that CC1A behaved as a competitor of toxofilin for the binding to G-actin suggesting that the toxofilin domain that binds G-actin is located in the CC1A domain.
We then tested whether the G-actin-binding property of CC1A was associated with a sequestering activity already described for the full-length toxofilin. To this end, we performed pyrene actin assays which demonstrated that both CC1 and CC1A polypeptides were sufficient to elicit a sequestering effect on G-actin (Figure 1E). The lower sequestering activity exhibited by CC1A in comparison with CC1 and assessed by the significant differences observed in steady-state values using the Student's t test (22.7 arbitrary units for CC1 and 62.8 arbitrary units for CC1A, representative of triplicates for three different experiments) is likely to result from a difference in CC1A and CC1 affinity to G-actin. In control, as expected from the pull-down assays, GST–CC2 did not modify the kinetics of actin polymerization (Figure 1F).
Three linear peptidic sequences within CC1A bind to G-actin
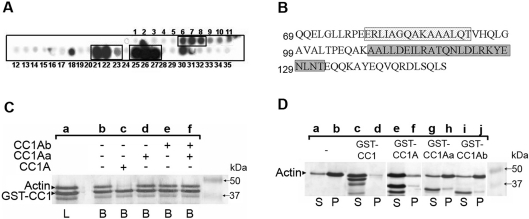
To identify the important amino acid residues of the CC1A polypeptide that could account for G-actin binding, we performed a peptide-spot assay using synthetic overlapping dodecapeptides spotted onto a cellulose membrane. The entire sequence of CC1A was represented, and each peptide differed from the following by a shift of two amino acids. When probed with biotinylated G-actin as described in the Experimental section, three series of spots were reproducibly detected (repeated 3 times) (Figure 2A). As seen in Figure 2(B), the spots numbered from 6 to 8 sharing the ‘AGQAKAAA’ sequence, bound to G-actin under the present experimental conditions. The spots 21–23 sharing the ‘DEILRATQ’ sequence and the spots 25–27 having in common the ‘NLDLRKYE’ sequence reacted the most strongly with G-actin (Figure 2A and 2B). Screening Uniprot/Swiss-Prot protein databases (216380 sequences May 2006) using the ‘pattern’ [19] and ‘pattinprot’ programs [20] identified a number of putative or well-characterized actin-interacting proteins that displayed high similarity (>70%) to the degenerated ‘AGQA[K/R]AAA’ (Table 1A) and ‘DEIL[R/K]A[T/S]Q’ motifs (Table 1B). For example, angiomotin, several mammalian myosins and talins were detected by the ‘AGQAKAAA’ motif. Several mammalian scinderins (also called adseverin) and mammalian actin-depolymerizing factors, which are all known to bind to G-actin, were identified by the ‘DEILRATQ’ motif. For this latter motif, myosin type II heavy chain, myosin Vc, skeletal muscle troponin C, neural spinophilin (neurabin-II) and the nuclear anchorage protein nesprin were also identified (Table 1B). In contrast, the third CC1A G-actin-binding motif identified only one known actin-binding protein in the database suggesting that the ‘NLDL[R/K][R/K]YE’ sequence is a more toxofilin-specific G-actin-binding motif. It is the microtubule–actin crosslinker MACF1 belonging to the spectraplakin family of cytoskeletal crosslinking proteins [21].
Figure 2. Toxofilin sequesters G-actin via the CC1A domain that contains three binding sites.
(A) The entire CC1A amino acid sequence was represented on a cellulose membrane by successive spots of dodecapeptides shifted by two amino acids and then probed with biotinylated G-actin and HRP-conjugated streptavidin. (B) The sequences corresponding to positive spots using chemiluminescence (A) are presented and framed. The first amino acid of each lane is numbered on the left. (C) Actin-binding competition assays were performed on immobilized GST–CC1 using G-actin preincubated with CC1A (lanes a–c), with CC1Aa (lane d) or CC1Ab (lane e) or with a mix of CC1Aa and CC1Ab (lane f). Samples were treated as described for Figure 1. L indicates the total load and B indicates the total bound fraction. The presence of the competitive fragment is indicated above the gel photograph. Arrowheads on the left indicate the position of G-actin and GST–CC1 on the gel. (D) Actin polymerization assays were performed by adding 100 mM KCl, 2 mM MgCl2 and 1 mM Na2ATP to G-actin overnight at 4 °C and further centrifugation. Supernatants (S) and pellets (P) were subjected to SDS/PAGE followed by Coomassie Blue staining. Actin alone polymerized efficiently (lanes a–b) whereas G-actin incubated with GST–CC1 (lanes c–d) and GST–CC1A (lanes e–f) did not. In contrast, GST–CC1Aa (lanes g–h) and GST–CC1Ab (lanes i–j) failed to prevent actin polymerization.
Table 1. ‘Pattinprot’ screening analysis of the UniProt/Swiss-Prot database with two toxofilin actin-binding motifs: (a) ‘AGQA[K/R]AAA’ and (b) ‘DEIL[R/K]A[T/S]Q’.
| (a) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Primary accession number | Protein name | Sequence | |||||||
| A | G | Q | A | K/R | A | A | A | ||
| CAB72264 | Toxofilin | A | G | Q | A | K | A | A | A |
| Q4VCS5 | Angiomotin | A | G | Q | i | p | A | A | A |
| Q21624 | Coronin-like | A | G | Q | r | R | A | A | A |
| Q9QY06 | Myosin-9B | A | e | Q | A | R | e | A | A |
| P05659 | Myosin-2 heavy | A | q | e | A | R | A | A | A |
| O96064 | Paramyosin | A | G | l | A | K | A | k | A |
| P54939 | Talin-1 chicken | A | q | Q | A | K | p | A | A |
| P26039 | Talin-1 mouse | A | s | Q | A | K | p | A | A |
| Q9Y4G6 | Talin-2 human | A | k | Q | A | a | A | A | A |
| Q7ILX4 | Talin-2 mouse | A | k | Q | A | a | A | A | A |
| (b) | |||||||||
| Primary accession number | Protein name | Sequence | |||||||
| D | E | I | L | R/K | A | T/S | Q | ||
| CAB72264 | Toxofilin | D | E | I | L | R | A | T | Q |
| Q28046 | Adseverin bovine | y | E | r | L | K | A | S | Q |
| Q60604 | Adseverin mouse | y | E | r | L | K | A | S | Q |
| Q29297 | Adseverin pig | y | E | r | L | K | A | S | Q |
| Q28372 | ADF horse | f | E | r | L | K | A | T | Q |
| P06396 | ADF human | y | E | r | L | K | A | T | Q |
| P13020 | ADF mouse | f | E | f | L | K | A | T | Q |
| P20305 | ADF pig | y | E | r | L | K | A | T | Q |
| Q96SB3 | Neurabin II human | D | E | h | L | R | e | T | Q |
| Q6R891 | Neurabin II mouse | D | E | h | L | R | e | T | Q |
| O35274 | Neurabin II rat | D | E | h | L | R | e | T | Q |
| Q8WXH0 | Nesprin 2 | D | E | d | L | s | A | T | Q |
| P02588 | Troponin chicken | g | E | I | L | R | A | T | g |
| P10246 | Troponin melga | g | E | I | L | R | A | T | g |
| Q9NQX4 | Myosin 5C | v | E | I | L | R | A | S | k |
Based on these results, we performed competitive assays for binding to G-actin using two polypeptides from CC1A, named CC1Aa (Gln69–Thr119) and CC1Ab (Gln120–Ser149) with GST–CC1A. As observed in Figure 2(C), addition of CC1A (lanes b and c) but not the addition of CC1Aa or CC1Ab (Figure 2C, lanes d–f) efficiently competed with GST–CC1 for binding to G-actin. Interestingly, a mixture of CC1Aa and CC1Ab could not restore the competitive property of CC1A, suggesting that the affinities of the two independent peptides for G-actin were lower than the affinity of the CC1A fragment and that the integrity of the sequence was required for binding to G-actin. Using an actin polymerization pull-down assay, we also tested whether CC1Aa and CC1Ab could prevent the polymerization of actin. As seen in Figure 2(D), the integrity of CC1A was required to fulfil the toxofilin-sequestering property.
NoCC binds to G-actin but does not sequester or nucleate G-actin
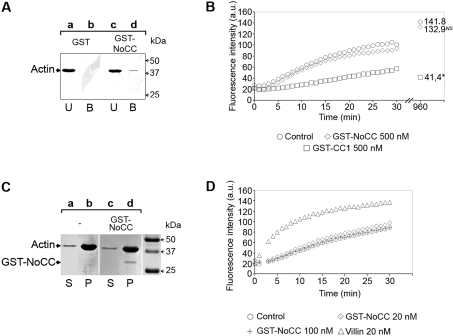
As presented above (Figure 1B), the 7.5% polyacrylamide native gel assay suggested that incubation of NoCC with G-actin partially transformed actin monomers into oligomers too large to enter the gel. Additionally, NoCC was found to bind to G-actin in a peptide-spot assay (results not shown). Therefore we investigated whether NoCC could capture G-actin in pull-down assays (n=6). As assessed by Western blotting (Figure 3A), under the present experimental conditions, GST–NoCC captured a small but reproducible amount of G-actin (lanes c and d) whereas GST (lanes a and b) or GST–CC2 (results not shown) did not. Moreover, in agreement with the native gel data, we found that GST–NoCC did not significantly affect the kinetics of actin polymerization upon addition of salt, magnesium and ATP in a pyrene actin assay (Figure 3B). This result indicated that, at identical concentrations, GST–NoCC did not have the sequestering activity of CC1 (Figure 3B). As expected, steady-state values for the sequestering CC1 were statistically different from those obtained with both NoCC and control. However, we reproducibly noticed that the steady-state values of fluorescence intensity associated with the amount of F-actin were slightly lower upon addition of GST–NoCC. The samples were subjected to ultracentrifugation and gel electrophoresis. As shown in Figure 3(C), the amounts of F-actin were similar whether GST–NoCC was added or not.
Figure 3. NoCC binds to G-actin and does not affect the rate and the amount of polymerization.
(A) Actin binding assays were carried out using immobilized GST (lanes a and b) or immobilized GST–NoCC (lanes c and d) and samples were analysed by Western blotting using anti-actin antibodies (clone C4). (B) Actin polymerization kinetics in the presence or absence of 500 nM GST–NoCC (◇) or 500 nM GST–CC1 (□) polypeptides using pyrene actin. Numbers on the right of the curves represent the actin steady-state values obtained 16 h after the assay. *The differences with the control values (○) are statistically significant at P=0.05 (Student's t test). NS, not significant; a.u., arbitrary units. (C) Actin polymerization assays were performed as described for Figure 2. Actin alone under polymerizing conditions (lanes a and b) and actin incubated with GST–NoCC in the same conditions (lanes c and d). Supernatants (S) and pellets (P) were subjected to SDS/PAGE and further Coomassie Blue staining. For each panel, the positions of actin and GST–NoCC are indicated with arrowheads on the left. (D) Actin nucleation assay in the presence or absence of 20 nM (◇) or 100 nM GST–NoCC (+) polypeptides using pyrene actin. As a positive control, villin was used at 20 nM (△). a.u., arbitrary units.
Since GST–NoCC weakly captured G-actin, using the same pyrene actin assay, we investigated whether it could induce actin nucleation. Although the well-known actin-binding protein villin displayed a clear nucleating activity at 20 nM in the present experimental conditions, GST–NoCC was unable to do so at both 20 and 100 nM (Figure 3D). Finally, we further tested whether the Ser53 carried by NoCC, and known to modulate toxofilin affinity to G-actin through phosphorylation/dephosphorylation, had any effect on actin nucleation or on the extent of actin polymerization and found no significant effect (results not shown).
The CC1–G-actin complex binds to PP2C through CC1B but not through CC1A
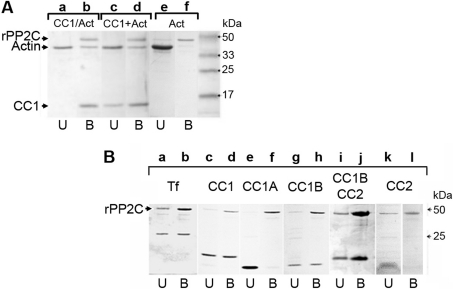
We previously showed that PP2C binds the actin–toxofilin complex [15]. To define the interacting domains between the three partners, toxofilin, actin and PP2C, we performed a pull-down assay using purified recombinant parasite PP2C, rPP2C. rPP2C was pre-immobilized on Ni-NTA–agarose through its histidine tag and then incubated with or without toxofilin polypeptides and G-actin. We first tested whether the CC1 domain of toxofilin was sufficient to bind to both G-actin and to PP2C. As shown in Figure 4(A), the tripartite complex was formed in vitro using the CC1 domain. This occurred upon incubation of rPP2C with the preformed CC1–G-actin complex (CC1–G-actin) (Figure 4A, lane b) as well as upon sequential incubation of rPP2C with CC1 and then G-actin (CC1+G-actin) (Figure 4A, lane d) provided that magnesium was added to the mix in the latter situation. Since G-actin did not bind directly to PP2C under the present experimental conditions (Figure 4A, lane f), these data strongly suggested that toxofilin CC1 contains a PP2C-binding site distinct from the actin-binding sites. Of note, the presence of PP2C did not affect the sequestering effect of CC1 in our actin polymerization assay (results not shown).
Figure 4. The CC1–G-actin complex binds to PP2C through CC1B but not through CC1A.
(A) Immobilized rPP2C on Ni-NTA was incubated with either the preformed CC1–G-actin complex (lanes a and b) or sequentially with CC1 then G-actin (lanes c and d) or with G-actin alone (lanes e and f) for 1 h at 4 °C. After extensive washings, Ni-NTA resin-bound proteins were eluted in SDS sample buffer and subjected to SDS/PAGE and Coomassie Blue staining. U indicates the total unbound fraction and B indicates the total bound fraction. Arrowheads on the left indicate the positions of G-actin, CC1 and rPP2C. (B) Immobilized rPP2C was incubated with toxofilin (Tf; lanes a and b), CC1 (lanes c and d), CC1A (lanes e and f), CC1B (lanes g and h), CC1BCC2 (lanes i and j) or CC2 (lanes k and l). Samples were treated as described for Figure 4(A). The arrowhead on the left indicates the position of rPP2C.
To define better the interaction between toxofilin and PP2C, we incubated rPP2C with full-length or truncates of toxofilin. While full-length toxofilin and CC1 were trapped by rPP2C as expected (Figure 4B, lanes b and d), only polypeptides carrying the CC1B sequence (i.e. CC1B and CC1BCC2) interacted with rPP2C (Figure 4B, lanes h and j). CC1A or CC2 polypeptides that lack the CC1B sequence did not bind to rPP2C (Figure 4B, lanes f and l). Similar results were obtained when we used the vertebrate purified recombinant Xenopus PP2C (results not shown).
Toxofilin displays two potential binding sites for PP2C within CC1B and NoCC respectively
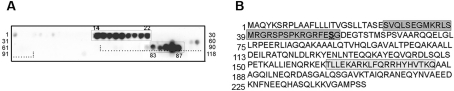
To identify the important amino acid residues of toxofilin that could account for rPP2C binding, we performed a peptide-spot assay on a nitrocellulose membrane spotted with synthetic overlapping dodecapeptides shifted by two amino acids that covered the entire toxofilin sequence. Membranes were probed with 4 μg/ml of purified rPP2C as described in the Experimental section. Two series of spots were reproducibly detected. One containing the spots 83–87 localized within the CC1B domain (Figure 5A, dotted lines) consistent with the binding assay. Interestingly, the last three spots (85–87) displayed a stronger signal than the two previous ones (83–84) (Figure 5A) and corresponded to the ‘KARKLFQRRHYHVTKQ’ sequence (Figure 5B). Since no binding was observed for the CC1A-corresponding peptide spots, these data again agree with our binding assay (Figure 4B) that favours an interaction site for PP2C within the CC1B region of CC1. Besides CC1B, PP2C also bound strongly to a 28 amino acid long sequence comprised within the spots 14–22 ‘SVQLSEGMKRLSMRGRSPSPKRGRFESG’ (Figure 5B). Interestingly, the signal intensity associated with spots 14–18 was stronger than the signal intensity observed for spots 19–22 (Figure 5A). Since spot 14 (SVQLSEGMKRLS) did not share any amino acid with the spots 21 and 22, it is likely that NoCC carries two independent but connected binding sites for PP2C. In addition, this second binding site within NoCC displays at its C-terminal part the Ser53 that we have previously shown to be phosphorylated by caseine kinase II and a substrate of PP2C.
Figure 5. Two toxofilin amino acid sequences bind to rPP2C using peptide-spot mapping.
(A) The entire toxofilin amino acid sequence was represented on a cellulose membrane by spots of dodecapeptides shifted by two amino acids. The CC1B region is represented with dotted lines. The membrane was successively probed with rPP2C, polyclonal anti-PP2C antibodies and HRP-conjugated anti-rabbit antibodies. (B) The sequences corresponding to positive spots using chemiluminescence (A) are presented and framed and the first amino acid of each lane is numbered on the left. In the first series 28 amino acid positive sequence, the known PP2C substrate Ser53 is shown in bold and underlined.
DISCUSSION
The 27 kDa toxofilin has been characterized as a G- and F-actin-binding protein in vitro using mammalian muscle actin and in heterologous systems such as mammalian cells. It was also found to tightly bind to parasite G-actin [14]. Phosphorylation of toxofilin on Ser53 by casein kinase II decreases its affinity for G-actin by 14-fold, and a type 2C phosphatase selectively dephosphorylates Ser53, thereby stabilizing the actin–toxofilin complex [15]. It is still unclear whether toxofilin targets parasite or mammalian actin or even both. Recent data from a rhoptry proteome analysis identified toxofilin as a rhoptry protein [16]. As such, toxofilin could be secreted from the secretory rhoptry organelle during host cell invasion, as has been shown for other rhoptry proteins which can remain in the vacuolar space or eventually be inserted into the parasitophorous membrane [22–24]. If toxofilin is released in the host cell cytoplasm, it is likely to act on the host cell actin to facilitate parasite progression through the host cell subcortical actin. In the present study, we used mammalian actin to undertake a structure–function analysis of toxofilin. We characterized toxofilin domains and amino acid sequences that account for the G-actin and PP2C binding properties.
Using native gel electrophoresis and pull-down assays, we demonstrated that toxofilin carries a domain that is sufficient for binding to G-actin and to elicit sequestration of actin monomers in an actin polymerization assay. We called this domain CC1A and it is located within a region of 9 kDa that encompasses the first coil–coiled domain. Using a peptide-spot mapping assay, we detected three series of overlapping peptides in CC1A that bind to G-actin. Screening of UniProt/Swiss-Prot protein data bases (216380 sequences May 2006) using ‘pattinprot’ with the common motif of these different overlapping peptides ‘AGQA[K/R]AAA’ provided more than 1000 hits (n=1054) when a 70% stringency threshold of similarity was applied, among which at least nine are well-known actin-binding proteins. This does not mean that such a sequence should be considered as a signature motif that accounts for the binding property to actin. It could however provide an indication for further and more accurate studies on proteins for which a putative binding property to actin has never been investigated so far or for proteins already reported to bind to actin. Peptide spot assays on peptides carrying systematic mutations were performed and the results prompted us to produce CC1 polypeptides into which we introduced single (Leu81, Lys87 and Leu91), double and triple mutations in the ‘ERLIAGQAKAAALQTVHQ’ motif but none of these mutations prevented actin binding to CC1 in pull-down assays suggesting that other sites might be sufficient to capture G-actin (results not shown). By contrast, disrupting the ‘DEILRATQNLDLRKYENL’ in the middle of the sequence after Thr119 had a major effect on the actin-binding properties of CC1A, suggesting that this sequence represents a crucial site for actin binding. When this ‘DEIL[R/K]A[T/S]Q’ degenerated motif was analysed in silico, 487 sequences were identified among which were 14 well-characterized G- and/or F-actin-binding proteins such as members of the ADF/cofilin family, the gelsolin and adseverin families and the myosin superfamily. This analysis supports our experimental data with toxofilin. On the other hand, when the ‘NLDL[R/K][K/R]YENL’ motif was examined in silico, only one of the 400 proteins detected was indeed described as interacting with actin but also with microtubules [25] and possibly with intermediate filaments as well. It is the microtubule–actin crosslinker MACF1. It is tempting to speculate that toxofilin may act in regulating microtubule and actin cytoskeleton interactions to support the development of the vacuole throughout the intracellular life of T. gondii. The parasitophorous vacuole is known to rapidly relocate towards the nucleus after invasion in a microtubule-dependent process.
We also found additional regions of toxofilin with actin-binding properties, in particular the NoCC domain. In a native gel assay, NoCC addition did not induce a similar shift of G-actin as to CC1 but rather prevented actin from running into the gel probably as a result of larger complexes between NoCC and several actin molecules. However, NoCC did not display the sequestering effect elicited by CC1 or any detectable nucleating activity in the pyrene actin assay. These data strongly suggest that apart from CC1A, toxofilin exhibits additional actin-binding sites but with different binding features and properties. The complete resolution of the three-dimensional structure and a thermodynamic analysis would provide answers to these questions. Based on such three-dimensional data, it would be informative to introduce mutations and to assay in vivo whether they have an impact on the parasite survival.
Since we previously reported that phosphorylation contributes to the toxofilin binding properties onto actin, and that PP2C co-purified in the actin–toxofilin complex, we also mapped the interactive regions of toxofilin with PP2C. In the present study, we show that the toxofilin region encoding the first coil–coiled domain, and closely linked to the C-terminal part of CC1A, bound to PP2C, as supported by both pull-down and peptide-spot mapping assays. An additional sequence in NoCC that bound to PP2C could represent two distinct but connected binding sites according to the peptide-spot assay. This sequence in NoCC also contains the amino acid Ser53 previously shown to be a substrate of PP2C. When non-redundant protein databases corresponding to more than 3000000 sequences were screened with the ‘LFQRRHYHVT’ motif and imposing a 85% stringency threshold in similarity, only toxofilin was pulled out whereas 17 hits were found when the stringency threshold for similarity was decreased to 70%. Among these positive hits, none corresponded to a protein yet assigned as a PP2C partner. When the same screen was applied with a degenerated sequence for basic and alkyl phosphorylatable residues ‘LFQ[R/K][R/K]HYHV[T/S]’, we obtained only toxofilin at a 90% threshold and 34 hits at a 70% threshold of similarity. Unlike for many kinases or for the PP1 and PP2A serine/threonine phosphatases, there are no clear sequence ‘signatures’ for PP2C dephosphorylation targets although the presence of a basic N-terminal residue at position- 3 and no proline residue adjacent to the C-terminal part of the phosphorylation have been shown to favour dephosphorylation by PP2C [26]. Toxofilin carries an arginine residue at position 50 and the absence of a proline residue after the Ser53 up to position 60 which is consistent with the theoretical favourable motif for being a PP2C substrate.
Characterizing PP2C substrates is hampered by the lack of specific inhibitors or activators, although several major targets have already been identified. These are mainly related to the cell cycle [27,28] or to cell apoptosis with the Bcl-2 family pro-apoptotic member BAD [29]. Mammalian PP2Cs are also increasingly analysed for their role in the homoeostasis of the central nervous system as for example PP2Cα interacts with calcium channels and contributes to synaptic transmission in neurons [30]. Therefore the dissection of a defining substrate such as toxofilin for the amino acid requirements that promote both PP2C binding and activity is among the first to be reported and should provide a useful reagent for both PP2C enzymatic characterization and broad modulatory molecule screening. In the future, in vivo strategies aiming at competing with endogenous PP2C may rely on non human-related sequences such as the toxofilin identified binding/substrate polypeptide.
Acknowledgments
We thank Dr R. Ménard (Institut Pasteur, Paris, France) for helpful critical reading of the manuscript. We are grateful to Dr Fernando Rocal (Centro Nacional de Biotecnologia, Departamento de proteomica, UAM, Madrid, Spain) for providing us with the membrane spot material. This work was funded by the Centre National de la Recherche Scientifique (ATIP grant to I.T.) and by the Ministry of Research (ACI Microbiology grant to I.T. and X.C., number AMA03042KSA). G.J. was supported by the Centre National de la Recherche Scientifique with a PhD fellowship (BDI, # NL/05/003).
References
- 1.Morisaki J. H., Heuser J. E., Sibley L. D. Invasion of Toxoplasma gondii occurs by active penetration of the host cell. J. Cell Sci. 1995;108:2457–2464. doi: 10.1242/jcs.108.6.2457. [DOI] [PubMed] [Google Scholar]
- 2.Sinai A. P., Webster P., Joiner K. A. Association of host cell endoplasmic reticulum and mitochondria with the Toxoplasma gondii parasitophorous vacuole membrane: a high affinity interaction. J. Cell Sci. 1997;110:2117–2128. doi: 10.1242/jcs.110.17.2117. [DOI] [PubMed] [Google Scholar]
- 3.Black M. W., Boothroyd J. C. Lytic cycle of Toxoplasma gondii. Microbiol. Mol. Biol. Rev. 2000;64:607–623. doi: 10.1128/mmbr.64.3.607-623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moudy R., Manning T. J., Beckers C. J. The loss of cytoplasmic potassium upon host cell breakdown triggers egress of Toxoplasma gondii. J. Biol. Chem. 2001;276:41492–41501. doi: 10.1074/jbc.M106154200. [DOI] [PubMed] [Google Scholar]
- 5.Hoff E. F., Carruthers V. B. Is Toxoplasma egress the first step in invasion? Trends Parasitol. 2002;18:251–255. doi: 10.1016/s1471-4922(02)02240-7. [DOI] [PubMed] [Google Scholar]
- 6.Hill D., Dubey J. P. Toxoplasma gondii: transmission, diagnosis and prevention. Clin. Microbiol. Infect. 2002;8:634–640. doi: 10.1046/j.1469-0691.2002.00485.x. [DOI] [PubMed] [Google Scholar]
- 7.Montoya J. G., Liesenfeld O. Toxoplasmosis. Lancet. 2004;363:1965–1976. doi: 10.1016/S0140-6736(04)16412-X. [DOI] [PubMed] [Google Scholar]
- 8.Dobrowolski J. M., Sibley L. D. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell. 1996;84:933–939. doi: 10.1016/s0092-8674(00)81071-5. [DOI] [PubMed] [Google Scholar]
- 9.Dobrowolski J. M., Carruthers V. B., Sibley L. D. Participation of myosin in gliding motility and host cell invasion by Toxoplasma gondii. Mol. Microbiol. 1997;26:163–173. doi: 10.1046/j.1365-2958.1997.5671913.x. [DOI] [PubMed] [Google Scholar]
- 10.Meissner M., Schluter D., Soldati D. Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science. 2002;298:837–840. doi: 10.1126/science.1074553. [DOI] [PubMed] [Google Scholar]
- 11.Carruthers V. B., Giddings O. K., Sibley L. D. Secretion of micronemal proteins is associated with Toxoplasma invasion of host cells. Cell. Microbiol. 1999;1:225–235. doi: 10.1046/j.1462-5822.1999.00023.x. [DOI] [PubMed] [Google Scholar]
- 12.Soldati D., Dubremetz J. F., Lebrun M. Microneme proteins: structural and functional requirements to promote adhesion and invasion by the apicomplexan parasite Toxoplasma gondii. Int. J. Parasitol. 2001;31:1293–1302. doi: 10.1016/s0020-7519(01)00257-0. [DOI] [PubMed] [Google Scholar]
- 13.Jewett T. J., Sibley L. D. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol. Cell. 2003;11:885–894. doi: 10.1016/s1097-2765(03)00113-8. [DOI] [PubMed] [Google Scholar]
- 14.Poupel O., Boleti H., Axisa S., Couture-Tosi E., Tardieux I. Toxofilin, a novel actin-binding protein from Toxoplasma gondii, sequesters actin monomers and caps actin filaments. Mol. Biol. Cell. 2000;11:355–368. doi: 10.1091/mbc.11.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delorme V., Cayla X., Faure G., Garcia A., Tardieux I. Actin dynamics is controlled by a casein kinase II and phosphatase 2C interplay on Toxoplasma gondii toxofilin. Mol. Biol. Cell. 2003;14:1900–1912. doi: 10.1091/mbc.E02-08-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradley P. J., Ward C., Cheng S. J., Alexander D. L., Coller S., Coombs G. H., Dunn J. D., Ferguson D. J., Sanderson S. J., Wastling J. M., Boothroyd J. C. Proteomic analysis of rhoptry organelles reveals many novel constituents for host–parasite interactions in Toxoplasma gondii. J. Biol. Chem. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- 17.Pardee J. D., Spudich J. A. Purification of muscle actin. Methods Cell Biol. 1982;24:271–289. doi: 10.1016/s0091-679x(08)60661-5. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Cockwell K. Y., Giles I. G. Software tools for motif and pattern scanning: program descriptions including a universal sequence reading algorithm. Comput. Appl. Biosci. 1989;5:227–232. doi: 10.1093/bioinformatics/5.3.227. [DOI] [PubMed] [Google Scholar]
- 20.Combet C., Blanchet C., Geourjon C., Deleage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 21.Kodama A., Karakesisoglou I., Wong E., Vaezi A., Fuchs E. ACF7: an essential integrator of microtubule dynamics. Cell. 2003;115:343–354. doi: 10.1016/s0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- 22.Beckers C. J., Dubremetz J. F., Mercereau-Puijalon O., Joiner K. A. The Toxoplasma gondii rhoptry protein ROP 2 is inserted into the parasitophorous vacuole membrane, surrounding the intracellular parasite, and is exposed to the host cell cytoplasm. J. Cell Biol. 1994;127:947–961. doi: 10.1083/jcb.127.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joiner K. A., Bermudes D., Sinai A., Qi H., Polotsky V., Beckers C. J. Structure and function of the Toxoplasma gondii vacuole. Ann. N.Y. Acad. Sci. 1996;797:1–7. doi: 10.1111/j.1749-6632.1996.tb52943.x. [DOI] [PubMed] [Google Scholar]
- 24.Alexander D. L., Mital J., Ward G. E., Bradley P., Boothroyd J. C. Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS Pathog. 2005;1:e17. doi: 10.1371/journal.ppat.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun D., Leung C. L., Liem R. K. Characterization of the microtubule binding domain of microtubule actin crosslinking factor (MACF): identification of a novel group of microtubule associated proteins. J. Cell Sci. 2001;114:161–172. doi: 10.1242/jcs.114.1.161. [DOI] [PubMed] [Google Scholar]
- 26.Pinna L. A., Donella-Deana A. Phosphorylated synthetic peptides as tools for studying protein phosphatases. Biochim. Biophys. Acta. 1994;1222:415–431. doi: 10.1016/0167-4889(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 27.De Smedt V., Poulhe R., Cayla X., Dessauge F., Karaiskou A., Jessus C., Ozon R. Thr-161 phosphorylation of monomeric Cdc2: regulation by protein phosphatase 2C in Xenopus oocytes. J. Biol. Chem. 2002;277:28592–28600. doi: 10.1074/jbc.M202742200. [DOI] [PubMed] [Google Scholar]
- 28.Cheng A., Ross K. E., Kaldis P., Solomon M. J. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 1999;13:2946–2957. doi: 10.1101/gad.13.22.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klumpp S., Selke D., Krieglstein J. Protein phosphatase type 2C dephosphorylates BAD. Neurochem. Int. 2003;42:555–560. doi: 10.1016/s0197-0186(02)00174-2. [DOI] [PubMed] [Google Scholar]
- 30.Li D., Wang F., Lai M., Chen Y., Zhang J. F. A protein phosphatase 2Cα-Ca2+ channel complex for dephosphorylation of neuronal Ca2+ channels phosphorylated by protein kinase C. J. Neurosci. 2005;25:1914–1923. doi: 10.1523/JNEUROSCI.4790-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]