Abstract
The transport of the plant hormone auxin has been under intense investigation since its identification 80 years ago. Studies have gradually refined our understanding of the importance of auxin transport in many aspects of plant signalling and development, and the focus has intensified in recent years towards the identification of the proteins involved in auxin transport and their functional mechanism. Within the past 18 months, the field has progressed rapidly, with confirmation that several distinct classes of proteins, previously dubbed as ‘putative auxin permeases’ or ‘auxin transport facilitators’, are bona fide transporters of IAA (indol-3-ylacetic acid). In this review we will appraise the recent transport data and highlight likely future research directions, including the characterization of auxiliary proteins necessary for the regulation of auxin transporters.
Keywords: Arabidopsis thaliana (thale cress), ATP-binding- cassette transporter (ABC transporter), auxin permease (AUX1), gravitropism, membrane transport, pin-formed (PIN)
Abbreviations: AAAP, amino acid and auxin permease; ABC, ATP-binding cassette; AGR, from agravitropic; ARF, auxin-response transcription factor; AUX1, auxin permease; AVT, γ-aminobutyric acid/glycine vesicular transporter; AXR, auxin-resistant; 2,4-D, 2,4-dichlorophenoxyacetic acid; EIR, ethylene-insensitive root; ER, endoplasmic reticulum; EYFP, enhanced yellow fluorescent protein; GEF, guanine nucleotide-exchange factor; IAA, indol-3-ylacetic acid; IAAH, undissociated form of IAA; LAX, like AUX; MDR, multidrug resistance; MRP, multidrug-related protein; 1-NAA, naphth-1-ylacetic acid; NBD, nucleotide-binding domain; NPA, naphthylphthalamic acid; NPPB, 5-nitro-2-(3-phenylpropylamino)benzoic acid; PDR, pleiotropic drug resistance; PGP, P-glycoprotein; AtPGP, A. thaliana PGP; PIN, from pin-formed; TCDB, transporter classification database; TM, transmembrane; TMD, TM domain; TWD1, twisted dwarf 1
INTRODUCTION
The term ‘auxin’ is applied to a group of naturally occurring and synthetic indoles and related compounds, the most biologically abundant of which is IAA (indol-3-ylacetic acid), but which also includes indolebutyric acid, 4-chloroindol-3-ylacetic acid, phenylacetic acid, 1-NAA (naphth-1-ylacetic acid) and 2,4-D (2,4-dichlorophenoxyacetic acid), which are responsible for a diverse range of plant developmental processes [1]. Auxins are ubiquitous in plants and have a panoply of effects on plant development from the early stages of embryogenesis and influence vascular tissue differentiation, flowering and fruit development, root development, phototropism, gravitropism and senescence [2].
The majority of auxin's effects are elicited intracellularly, with at least some of these ascribed to auxin being a signalling molecule capable of causing rapid up-regulation of the transcription of genes whose promoter regions contain auxin-responsive elements. A mechanism describing the effects of auxin on the transcription of these genes has recently been proposed [3,4]. Briefly, the transcription of such genes is regulated by the balance of ARFs (auxin-response transcription factors) and a large family of auxin-induced gene products (the so-called ‘Aux/IAA’ proteins) that act as repressors of ARF function. Protein complex formation between ARF and Aux/IAAs leads to gene silencing [5]. However, the direct binding of auxin to an F-box protein {TIR1 (transport inhibitor response 1) [6]} enables formation of a multi-protein SCF (Skp1/Cullin/F-box) complex with Aux/IAA protein. The subsequent ubiquitination and proteasomal degradation of Aux/IAA frees the ARF, causing immediate transcription of auxin target genes [3,4]. Interestingly, among these are the Aux/IAA transcriptional repressors themselves, so establishing feedback regulation of the auxin signal. Feedback regulation of auxin recurs at the level of transport (see below).
Auxin is synthesized predominantly from tryptophan (although there are also tryptophan-independent synthetic routes) in young, growing regions of plants such as new leaf primordia [7]. A significant amount of auxin may also be stored as conjugates with sugars or amino acids [2]. Newly synthesized auxin, or auxin enzymatically liberated from storage forms, travels through a combination of two processes: (i) a rapid (5–10 cm/h) non-directional transport occurs in the vasculature that is augmented by (ii) polar auxin transport (5–10 mm/h), which establishes physiologically relevant auxin gradients [8]. Polar auxin transport, which is important in the early stage of seedling growth, appears to be superseded subsequently by phloem-based transport [9]. Within the shoot, polar auxin transport occurs in a ‘reverse fountain’ manner, with epidermal auxin streams directed towards the vascular tissues at leaf primordia [10,11]. In the root, auxin undergoes polar transport in two directions. There is both transport from the base (ground level) to the root apex (‘acropetal’ direction) through protophloem cells and movement in the opposite (‘basipetal’) direction through the lateral root cap (Figure 1), the asymmetry of this movement being responsible for generating the auxin gradients, mediating the gravitropic response (as described in the Cholodny–Went hypothesis [12–14]).
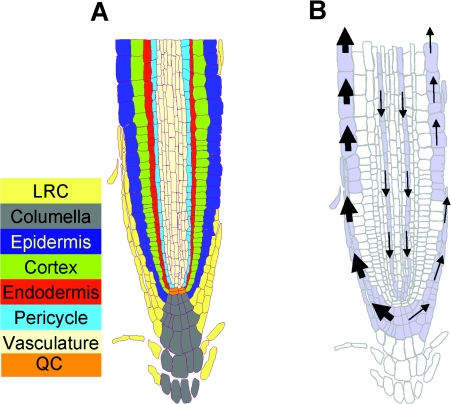
Figure 1. Auxin transport in roots.
(A) The cellular structure and different functional cell layers of the Arabidopsis root tip are shown. Abbreviations: QC, quiescent centre; LRC, lateral root cap. (B) Auxin is transported towards the root tip through protophloem cell files in the vasculature and then, in response to gravity, distributed asymmetrically by polarly localized auxin influx (AUX) and efflux (PIN) proteins in the lateral root cap, where the gravitropic response is effected. The thickness of arrows depicts the relative auxin flux.
The requirement to establish and maintain auxin distributions between and within tissues, and the need to import auxin as a signalling molecule, infers the requirement for both auxin-import and auxin-export proteins. A chemiosmotic hypothesis exists [15] to describe polar auxin transport in plant cells. As a weak acid (pKa 4.8) a significant fraction (at least 15%) of IAA will exist in the undissociated form (IAAH) at the pH of the plant extracellular apoplastic space (∼pH 5.5). As this uncharged IAAH is lipophilic, it would be expected to freely diffuse across the plasma membrane in the presence of a suitable concentration gradient, leading to the opinion that protein-driven auxin import may be of minor importance [16]. Notwithstanding this, early studies on auxin transport in plant cells in suspension culture revealed that the uptake rate was non-linear with respect to auxin concentration, suggesting that diffusion was not the only process. Furthermore, at a fixed external concentration of IAAH, there was a pH-dependence to auxin uptake, confirming the existence of a carrier-dependent contribution to the overall rate of import [15]. The relative contributions of diffusion and carrier-mediated import have recently been reviewed, leading the authors to conclude, on the basis of mathematical considerations and mutant studies, that the carrier-mediated uptake is the major contributor to polar auxin transport [14]. Once inside plant cells (pH 7–7.5), any uncharged IAAH will dissociate and exist as the anionic form, trapping auxin intracellularly. Therefore the existence of specific importers and exporters is required to effect continual polar flux.
IDENTIFICATION OF THE MAJOR PLAYERS IN PLANT INTERCELLULAR PROTEIN-MEDIATED AUXIN TRANSPORT
Identification of auxin-transport genes was facilitated by a combination of two main approaches: first, the wide availability of Arabidopsis mutants, isolated by forward and reverse genetics, with defective tropisms and stimulus–response coupling and, secondly, the altered sensitivity of these mutants to, or the potential to ‘phenocopy’ them with, synthetic auxin analogues or auxin-transport inhibitors [17]. For example, the agravitropic mutant Arabidopsis aux1 could be rescued with the freely-membrane-permeable auxin analogue 1-NAA, suggesting that the mutant was defective in auxin uptake [18]. Alternatively, the mutant PIN (an abbreviation of the mutant allele pin-formed, itself referring to the pin-shaped inflorescences occurring in the mutant plant) was phenocopied by the addition of the auxin-transport inhibitor NPA (naphthylphthalamic acid), leading to the cloning of an auxin- efflux carrier [19].
AUX/LAX (AUXIN PERMEASE/LIKE AUX) FAMILY
The first auxin permease gene, AUX1, was originally cloned a decade ago using a gene-tagging approach [20]. Whole genome sequencing projects have since identified four members in A. thaliana (thale cress) [21], five in Medicago truncatula (barrel medick) [22] and five in Oryza sativa (rice) [23,24]. Indeed, in all plant species investigated, there are sequence homologues of A. thaliana AUX1. Studies on Arabidopsis have shown that AUX1 is expressed in protophloem, columella, lateral root cap and expanding epidermal cells at the root apex, where it facilitates acropetal and basipetal auxin redistribution. In contrast, the three closely related genes termed LAX1, LAX2 and LAX3 are predominantly expressed in root and aerial vascular tissues [25]. The AUX/LAX putative auxin importers form a subgroup of the AAAP (amino acid and auxin permease) family identified by Saier and co-workers {classified as 2.A.18 in their TCDB (Transport Classification Database; http://www.tcdb.org/; [26]}. The general mechanism of transport remains to be elucidated, but is predicted to occur as proton symport, in concordance with the mechanism of the distantly related APC (amino acid–polyamine–organocation) superfamily. In non-plant species there are no well-documented sequence homologues (i.e. there are no non-plant members of 2.A.18.1), although distant sequence relationships have been documented with a proposed neutral-amino acid permease from the fungus Neurospora crassa and with two predicted open reading frames from the nematode worm Caenorhabditis elegans [26]. Of course, most organisms have amino acid permeases that are members of the AAAP family, but BLAST (Basic Local Alignment Search Tool) comparisons of these and AUX/LAX sequences suggest a very distant relationship [26]. For example, yeasts have at least seven presumed vacuolar amino acid permeases (AVT1-7; TCDB 2.A.18.5) which are less than 15% identical with AUX1 at the amino acid sequence level. Of interest to auxin-transporter biologists is the yeast mutant yap1-1 (in which the transcriptional activator Yap1 is absent), which shows heightened sensitivity to the effects of IAA on yeast (the compound induces invasive growth [27]) and shows increased transcription of AVT (γ-aminobutyric acid/glycine vesicular transporter) genes. Although deletion of any of the AVT genes in a yap1-1 background restores wild-type sensitivity to IAA, there is no direct auxin-transport-assay data on individual AVT proteins [27].
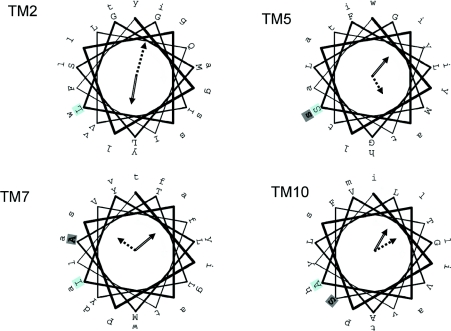
Typically AUX/LAX proteins consist of 460–490 amino acids with predicted molecular masses of ∼55 kDa. On SDS/PAGE, AUX1 migrates with an anomalously low molecular mass (∼45 kDa), which is unaffected by prior treatment with glycosidases, suggesting that there is little or no glycosylation, a finding in agreement with the sequence analysis, which indicated that there are no O-linked glycosylation sites and that the four putative N-linked glycosylation sites are predicted to be intracellular or membrane-embedded [28]. Multiple sequence alignment analysis of the AUX/LAX protein sequences reveals an extremely high degree of conservation over the entirety of the alignment (the majority of AUX/LAX proteins share in excess of 60% sequence identity), with the only exception being an ∼100-amino-acid-long insertion into the fourth predicted intracellular loop of the auxin-influx carrier-like protein 2 of the Momordica charantia (bitter gourd). Transmembrane topology prediction using multiple web-based applications [29] of any of the individual sequences produces data suggestive of between seven and 12 TM (transmembrane) helices. As with many other families of transporter, the shortness of several of the predicted intra- and extra-cellular loops hinders accurate prediction [30], although consensus analysis of the various predictions suggests the existence of 11 TM spans [28]. The predicted 11-TM topology requires full confirmation, but has received some support from experimental analysis of fusions of AUX1 with yellow fluorescent protein, which demonstrated that there is an odd number of TM spans, with the N-terminus being cytoplasmic [28]. In addition, and in agreement with the topological model, an affinity-tagged neutral amino acid transporter (a distant relative of the amino acid auxin permeases; TCDB 2.A.18.4) was shown to have an odd number (probably 11) TM spans when translated in vitro [31]. We have analysed the periodicity of the predicted TM spans of the multiple aligned AUX/LAX sequences (PERSCAN software analysis [32]) and this confirms that there is α-helical periodicity in, among other things, residue hydrophobicity and conservation. For example, TM2 has strong α-helical periodicity in residue conservation, indicating that there is a face of the predicted α-helix that is highly conserved (Figure 2). Similarly, there is high periodicity in residue hydrophobicity, with the hydrophobic face of the helix diametrically opposite the conserved face. For TM5, TM7 and TM10 there are also strong predictions of periodicity in residue conservation and hydrophobicity, with the identified faces either approximately shared (TM10) or removed by at least one helical turn (TM5 and TM7) (Figure 2). These data are not sufficient to predict a TM packing model for AUX1, and any functional significance awaits further mutational analysis, but it is of interest that three loss-of-function alleles of aux1, namely aux1-111, aux1-2 and aux1-118 [28], map to the centres of the hydrophilic face of TM5, the conserved face of TM7 and the hydrophilic face of TM10 respectively (residues in bold in Figure 2). With 11 predicted TMs it might seem incongruous to ask whether there is any evidence of internal sequence duplication in the AUX/LAX family. However, the ammonium transporters [exemplified by AmtB (ammonia transporter B) from Escherichia coli], which share an 11-TM helix motif and no internal sequence homology, display structural homology between helices 1 and 5 and helices 6 and 10 [33]. We note in this respect that TM5 and TM10 of the AUX/LAX family have hydrophilic faces containing serine residues, the mutation of which is associated with loss-of-function in AUX1 (Figure 2).
Figure 2. Periodicity analysis of the predicted TM α-helices of AUX/LAX proteins.
TM helices 2, 5, 7 and 10 of the AUX/LAX family have a periodicity in their predicted residue conservation and hydrophobicity. The linear sequence of each TM of A. thaliana AUX1 is projected on to a helical wheel, with the first amino acid of each helix set against a pale blue background. In each case the open arrow points to the centre of the hydrophobic face of the helix and the broken arrow to the centre of the conserved face of the helix. The length of the arrow is proportional to the strength of observed periodicity [32]. Loss-of-function missense mutations in AUX1 [28] in TM5, TM7 and TM10 are indicated in bold on a light background. Other predicted TM helices in AUX/LAX sequences show less convincing periodicity.
PIN FAMILY
The second family of putative auxin transporters to be identified was the PIN protein family. PIN1 was almost simultaneously cloned and sequenced as AGR [from the mutant allele agravitropic with a synonymous phenotype; also known as EIR (ethylene-insensitive root) and PIN2], and the two proteins were identified as probable auxin-efflux carriers and likely determinants of polar auxin transport in vascular [19] and root tissue [34–37] respectively. The extensive sequencing of plant genomes confirms the existence of multiple PIN isoforms in all species analysed, with eight in Arabidopsis [38] and Oryza [23,24] and ten in Medicago [22]. Localization studies demonstrate specific tissue distributions for several PIN family members. For example, PIN1 is predominantly found in the stele, whereas PIN2 is localized to the lateral root cap and epidermal and cortical cells [38].
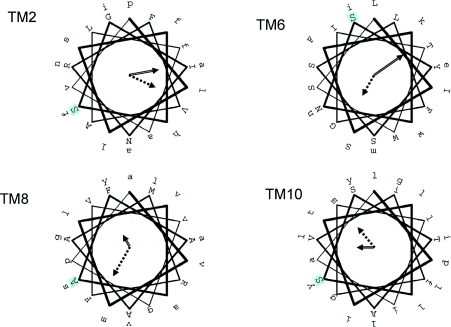
PIN proteins are classified into subfamily 2.A.69 (amino acid/polyamine/choline family of transporters) of the TCDB. Although DNA and protein sequence analysis is sufficient to define the existence of PIN subfamiles [38], there remains greater than 35% sequence identity between any pair of PIN proteins from higher plants. Full-length PIN sequences are ∼650 amino acids in length and have a predicted molecular mass of 65–70 kDa. The ‘denatured’ molecular mass by SDS/PAGE is close to 70 kDa [37], indicating that, for PIN2 at least, there is no substantial glycosylation, in agreement with predictions which would place all but one predicted N- and O-linked glycan sites in intracellular regions. TM topology predictions of PIN proteins are rather more consistent that those for AUX/LAX proteins, with ten spans being the consensus and most strongly predicted topology. If this prediction is correct (and there is no existing evidence to support this topology), then there are two blocks of five TM helices at either terminus, each of which is connected to the next by a short (at most 12 amino acids) inter-helical loop. Separating this ‘2×5’ TM motif is a long, presumably intracellular, loop of approx. 300 amino acids. Although there is no internal sequence homology between the two blocks of five predicted TM α-helices, there may well be structural homology between the two halves, as suggested above for the AUX/LAX family. As with AUX/LAX proteins, there is a significant α-helical periodicity to PIN TM sequences (Figure 3). PERSCAN analysis [39] identifies all TMs (except TM5 and TM7) as having periodicity in residue conservation and hydrophobicity. TMs 2 and 10 have a face of the helix that is both hydrophobic and conserved, whereas TM6 and TM8 have conserved faces that are distinct from the hydrophobic face. Again, the functional significance of this helical periodicity has not been investigated. Intriguingly, whereas there are no convincing homologues of AUX/LAX proteins in bacteria, there appear to be homologues of PIN proteins. Two small subfamilies of transporters, exemplified by MleP of the Oenococcus oeni [40] and MdcF of Klebsiella pneumoniae [41] (the former of which has some organic-acid-transport activity [40]) share the ten TM topology and share 20–25% identity with PIN proteins. However, MdcF and MleP lack the lengthy central hydrophilic domain. This domain is also absent in some plant PIN proteins (e.g. Arabidopsis PIN5) and shows much lower sequence similarity than the TM domains across the PIN family, and we suggest that this domain is involved in regulation via protein–protein interactions.
Figure 3. Periodicity analysis of the predicted TM α-helices of PIN proteins.
Several of the TM helices of the PIN family have periodicity; TM2, TM6, TM8 and TM10 only are shown for clarity. TM5 and TM7 show no periodicity. In each case the sequence of A. thaliana PIN1 is shown projected on to a helical wheel, with the first amino acid against a pale blue background. Arrow styles are as described in Figure 3.
MDR/PGP (MULTIDRUG RESISTANCE/P-GLYCOPROTEIN) FAMILY
The third group of membrane proteins implicated in auxin transport comprises members of the ubiquitous ABC (ATP-binding cassette) transporter family. ABC transporter sequences from A. thaliana have been subclassified into a number of distinct clades [42], broadly in line with the classification of yeast ABCs [43]. The two largest of these subfamilies in Arabidopsis are the MDR-type and the MRP (multidrug-related-protein)-type proteins so named because of their sequence homology with the well characterized broad-substrate-specificity drug pumps from mammalian systems [42,44]. These subfamily names are potentially misleading, as there are many examples of human ABC transporters that, by sequence homology, are classified as MDR-type ABC transporters (ABCB according to the nomenclature of [44]), but which play no role in drug transport. Indeed, expression of AtPGP1 (A. thaliana PGP; see below) in human HeLa cells does not result in enhanced transport of fluorescent substrates of human MDR1 (PGP), such as Rhodamine 123 and BODIPY® (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene)-vinblastine [45]. A similar caveat is appropriate for the MRP-type ABC transporters – the equivalent family in humans (ABCC transporters [44]) also contains proteins responsible for chloride-ion secretion [46] and regulation of an inwardly rectifying potassium channel [47].
The first association between auxin transport and ABC proteins was made following a screen for resistance to the anion-channel inhibitor NPPB [5-nitro-2-(3-phenylpropylamino)benzoic acid] [48]. One of the genes, overexpressed in NPPB-treated Brassica napus (oilseed rape) seedlings, was used to identify the Arabidopsis homologue, namely Atmdr1 (the gene nomenclature Atmdr1 is widespread in the literature, but the encoded protein is referred to as both AtMDR11 [42] or AtPGP19). In common with mammalian MDR-type ABC transporters [49], AtMDR1/AtMDR11/AtPGP19 and its closest sequence relative, AtPGP1 [42,50], are classified as full-length ABC transporters (with 1200–1300 amino acids), comprising two TMDs (TM domains), each of which is followed by a cytoplasmic NBD (nucleotide-binding domain) [51,52]. Structural studies of prokaryotic and eukaryotic ABC transporter NBDs have revealed that the domain adopts a highly conserved fold [53,54], and that co-ordination and subsequent hydrolysis of two ATP molecules by two interlocking NBDs is likely to represent a key conformational transition in the catalytic and transport cycles [55]. Structural data on the binding sites of transported substrates is more limited, but the majority of functional data on MDR-type ABC transporters [49] would support the hypothesis that auxin binds to the TMDs of AtPGPs.
The mutant phenotypes of Atmdr1 and a double mutant (Atmdr1.Atpgp1) are consistent with excess apical auxin, i.e. a failure to transport auxin basipetally from the shoot apex, whereas the Atpgp1 single-mutant phenotype is considerably more subtle [48,57]. A similar functional role for homologues of AtPGP1 in agronomically important dwarf maize (Zea mays) and Sorghum (sorghum) has recently been proposed [58]. More recently, AtPGP4 (which is localized to the root cap and elongation-zone epidermis) has been proposed to be involved in the auxin-controlled growth of lateral roots, although there are contradictions between the presented phenotypic data [59,60] regarding this effect, probably due to the use of different root growth media.
Among other Arabidopsis ABC transporters, an early study on the role of AtMRP5 in root development and stomatal opening was consistent with an auxin-transporter function [61], but more recent studies strongly suggest an alternative channel or channel-regulatory function for the protein [62]. Additionally, there is a suggestion that a plant peroxisomal ABC transporter, AtPxa1 (AtPmp2 [42]), is involved in transport of indol-3-ylbutyric acid into the peroxisome, where it undergoes a β-oxidation reaction to IAA [63]. Finally, a recent report identifies a homologue of the yeast pleiotropic drug-resistance subfamily of ABC proteins, namely AtPDR9 (A. thaliana pleiotropic drug resistance 9), as being responsible for 2,4-D resistance [64].
HETEROLOGOUS EXPRESSION OF AUX/LAX, PINs AND ATMDRs PROVIDES COMPELLING EVIDENCE FOR DIRECT ROLES IN IAA TRANSPORT
Most of the data supporting auxin transport by AUX/LAX, PINs and AtMDRs has, until recently, suffered from a major potential detraction. Studies of mutant plant alleles (or even transport in leaf protoplasts [45]) are open to other interpretations – for example, the Atmdr1 mutant plant shows clear alterations to auxin transport and auxin concentrations with phenotypic consequences, but it is well established that there are mammalian ABC proteins which are regulators of the activity of other proteins, rather than directly transporting themselves [65]. Such a caveat is noted by the authors of Atmdr1 studies [48,66]. Thus heterologous transport studies have been seen as necessary to rule out indirect effects. This requirement has led to the ambiguous label ‘putative auxin permeases’ and ‘auxin transport facilitators’ being applied to AUX1, PINs and AtPGPs.
Recently, however, a series of transport studies undertaken using heterologous expression of AUX/LAXs, PINs and AtPGPs in yeast, mammalian cells and microinjected Xenopus (South-African clawed frog) oocytes have provided concrete evidence that these proteins are all direct mediators of auxin transport. This shedding of the unwanted ‘putative’ tag now enables detailed characterization of the kinetics of auxin transport.
AUX/LAX TRANSPORT DATA
The most comprehensive analysis of a ‘putative’ auxin transporter has been provided from studies on Xenopus oocytes injected with transcripts encoding AUX1 [67]. Since the expression of proteins in single oocytes is undetectable by Western blotting, the Nielsen group microinjected RNA encoding EYFP (enhanced yellow fluorescent protein)-tagged AUX1, which is known to be able to rescue a mutant aux1 phenotype in plants [25]. EYFP–AUX1 was localized by confocal microscopy to the plasma membrane of oocytes [although correct targeting took 6 days compared with 2–3 days for many other transporters expressed in this system – an observation which may be of relevance, given the demonstrated dependence of AUX1 trafficking on AXR4 (auxin-resistant 4) described below], and uptake of IAA was demonstrated to be both pH- and IAA-concentration-dependent. The pH profile was consistent with a significant rate of diffusion of IAA (presumably as IAAH) below pH 5.5, but that, between pH 5.5 and pH 6.5, the net increase in IAA uptake (AUX1 injected versus uninjected controls) was most marked, in agreement with the pH optimum for active auxin uptake suggested 30 years ago [15]. At pH 6.4 (at which value over 95% of IAA will exist in the protonated form and thus diffusion will be highly restricted) the Km for IAA uptake was 800 nM, a value again consistent with earlier estimates for plant cells grown in suspension [15]. A number of excellent controls ensure that the observed uptake of [3H]IAA is AUX1-mediated. In particular, the authors demonstrated that three missense alleles of aux1, which in plants produce an agravitropic phenotype consistent with AUX1 misfunction [28], have an auxin-influx capacity which is either indistinguishable from that of the controls or is severely abrogated [67]. Importantly, all three mutant isoforms of AUX1 reached the plasma membrane, substantiating the notion that these alleles represent loss of function, rather than proteins with targeting defects. As the three point mutations are in predicted extracellular loops, it remains open to question how they affect AUX1 function [28]. Transport of [3H]IAA into oocytes was inhibited by unlabelled IAA and by 2,4-D, as well as by the auxin import inhibitors 1- and 2-napthoxyacetic acid [17,68], whereas addition of the auxin-efflux substrate 1-NAA and the auxin-efflux inhibitors NPA and 2,3,5-tri-iodobenzoic acid was without effect [67], suggesting that the substrate specificity of auxin transport through AUX1 is not dependent upon interactions with other membrane proteins. As discussed below, the same may not be true for PINs and AtMDR/PGP proteins.
PIN TRANSPORT DATA
Initial characterization of PIN2/AGR1/EIR1 expressed in baker's yeast (Saccharomyces cerevisiae) showed a small increase (15%) in IAA efflux from cells pre-loaded at pH 4.6 (a value at which 50% of IAA would exist in the uncharged IAAH form), when compared with the control (antisense PIN2) transformants. In addition, PIN2-transformed yeast was slightly more resistant to the toxic 5-fluoro-IAA, although this difference does not appear to be statistically significant [36]. A similar conclusion, based upon a disc assay test (rather than a radiolabelled transport assay), was obtained with PIN2 expressed in a strain of S. cerevisiae, which has enhanced susceptibility to fluorinated indoles [34].
A more detailed analysis of PIN-mediated auxin efflux has come recently with studies of multiple PIN proteins in several different cell lines [69]. The use of easily disrupted Arabidopsis cultured cells, more robust tobacco (Nicotiana tabacum) BY-2 cells [70], human HeLa cells and yeast were all required to provide convincing evidence that the export of auxins (typically measured by accumulation of [3H]auxins in the presence and absence of the auxin-efflux inhibitor NPA) was PIN-specific. High background efflux of auxins (due to endogenous proteins) was observed in both plant cell lines and in human HeLa cells [69]. The results obtained were consistent with PIN1, PIN2, PIN4, PIN6 and PIN7 being able to transport IAA, and the analogues 1-NAA and 2,4-D, which would be consistent with PIN in planta function. Reduced retention of 1-NAA by PIN3 has also been briefly examined in BY-2 cells as a surrogate marker for auxin efflux [71]. Of the non-plant expression systems employed [69], the sensitivity of transport assays seems much better in yeast cells than in HeLa cells, which may be a reflection of the yeast strain employed (JK93da; see below). However, the observed export of benzoic acid by PIN2 in yeast (and not HeLa cells) suggests that the substrate specificity in some heterologous expression systems may present inaccurately, as observed with AtMDRs ([69] and see below). Finally, the kinetics of export, revealed by wash-out experiments on yeast or tobacco BY-2 cells, show that efflux is rapid but saturable [69]. Despite these advances there are no data presented for the Km and Vmax of transport, and only limited data are presented for a non-functional mutant PIN2 isoform [69], in contrast with studies on AUX1 [67].
MDR/PGP TRANSPORT DATA
Initial studies on AtMDR11/AtPGP19 expressed in a S. cerevisiae strain deficient in several potential interfering ABC transporters (yeast PDR pumps; strain JK93da [72]) were unable to demonstrate auxin efflux [48], although there was increased binding of the auxin-transport inhibitor NPA to yeast cells, and both AtMDR11/AtPGP19 and AtPGP1 could be identified from NPA affinity chromatography of Arabidopsis plasma membranes [48]. In contrast, recent expression of the same protein in transformed tobacco BY-2 cells [69] and in vaccinia-virus-infected HeLa cells [73] has been shown to confer a reduced accumulation of [3H]NAA and [3H]IAA respectively in comparison with control cells. The difference between these findings and results obtained for yeast presumably lies in the altered targeting of the expressed protein in the cell lines. The close sequence homologue AtPGP1 has been shown to confer IAA-transport activity in yeast (S. cerevisiae JK93da) and reduced sensitivity to the IAA-sensitive strain yap1-1 [27,45], despite its much weaker phenotype in mutant plants. Confocal microscopy confirmed the plasma-membrane targeting of AtPGP1 in strain JK93da, and loading and efflux experiments at pH 4.6 showed evidence of greater IAA export in yeast transformed with AtPGP1 compared with untransformed controls [45]. Again there is no determination of Michaelis–Menten parameters for the AtMDRs, with much of the data being obtained from single IAA concentrations. Additionally, the substrate specificity of transport was unexpectedly broad (consistent with observations on AtPGP19 [69,73]), with a weak organic acid, benzoic acid, also showing enhanced transport in transformed cells [45] – in apparent contrast with previously obtained results showing failure of benzoic acid to prevent auxin binding to yeast expressing AtMDR11/AtPGP19 [48]. The same study presents additional data on AtPGP1 transfected into human HeLa cells, using vaccinia virus [45], with evidence that [3H]IAA is effluxed from expressing cells, which is partially inhibited by unlabelled IAA.
Interestingly, the inhibitor profile of AtPGP1-mediated auxin efflux was investigated by these authors. IAA efflux from JK93da yeast cells was inhibited by typical modulators of mammalian MDR pumps, including cyclosporin A and verapamil [74,75], yet AtPGP1 expressed in human HeLa cells did not transport typical MDR substrates such as Rhodamine 123 and vinblastine [45,76,77]. If AtPGP1 and AtPGP19 are acting as auxin-efflux carriers, then it is possible that substrate specificity is modulated by other proteins or the lipid environment. Alternatively, the relatively broad substrate specificity of human PGP [49] and other members of the human ABCB subfamily (e.g. [78]) leads to the suggestion that AtPGPs may be able to transport chemically distinct compounds such as benzoic acid. Further studies to determine the precise substrate specificity of AtPGP1/19 are required.
Transport of IAA by AtPGP4 has been investigated by two groups [59,60]. The latter group employed human Hela cells (essentially as described in [45]), whereas the former study employed the S. cerevisiae strains JK93da [72] and yap1-1 [27]. In JK93da yeast, expression of AtPGP4 increased sensitivity to 5-fluoro-IAA compared with control cells. Similar results were obtained with yap1-1 (which is sensitive to IAA) with hypersensitivity to IAA shown by AtPGP4 transformants. Both these findings argue for an import of IAA catalysed by AtPGP4 [59,60]. This would make AtPGP4 virtually unique amongst eukaryotic ABC transporters – although the one other published exception, MDR1 from Coptis japonica (Japanese goldthread), is a close sequence homologue of AtPGP4 [79]. However, the high degree of sequence similarity between AtPGP4 and the established export pump, human PGP (46% sequence identity over the entire length of the proteins), is difficult to reconcile with these findings.
The study by Terasaka's group [60] also attempted to show transport substrate (i.e. IAA) binding to AtPGP4 via an indirect assay, namely the ability of transport substrate to increase ‘vanadate-trapping’ of ABC transporters. For several ABC proteins it has been demonstrated that incubation of protein (e.g. in membranes) with enzymatic substrate (ATP) and vanadate traps the protein in a post-hydrolytic conformation in which ADP·vanadate is stably bound by the NBDs [80,81]. If photoactive 8-azido-[32P]ATP is substituted for ATP, then exposure of trapped protein to UV light results in cross-linking of ATP to protein and subsequent identification. Relying on the premise that transported substrates accelerate the rate of hydrolysis in ABC transporters (reviewed by [55]), it has been shown that transport substrates can also increase the extent of vanadate trapping [82]. For AtPGP4 there was increased vanadate trapping in the presence of IAA and NAA, but not other auxins, including 2,4-D [60]. However, direct drug-binding studies (which have been performed on numerous other ABC proteins [76,83,84]) have yet to be performed on any AtMDR, precluding definitive identification of substrate specificity.
CONTROL OF AUXIN-TRANSPORTER POLARITY AND FUNCTION
Recent investigations have identified several key mediators in the post-translational trafficking of auxin transporters. The auxin influx carrier AUX1 requires the AXR4 protein for correct subcellular localization [85]. AXR4 encodes a novel ER (endoplasmic reticulum)-localized protein [85]. Mutations in the AXR4 gene cause AUX1 to be retained in the ER of selected root tissues, resulting in the axr4 mutant displaying an aux1-like phenotype. It is currently unclear whether AXR4 directly interacts with AUX1 in the ER, where it may be required during the assembly of the multispan transporter. Alternatively, AXR4 may facilitate the trafficking of AUX1 [86]. Elucidation of AXR4 function awaits further experimentation.
Trafficking of selected PIN proteins has been demonstrated to be dependent on the GNOM protein [87]. The GNOM gene encodes a GDP/GTP exchange factor for small G-proteins of the ARF class {ARF-GEF (ARF-guanine nucleotide-exchange factor); [88,89]}. ARF-GEFs regulate cargo-selective vesicular trafficking [90]. GNOM is localized in endosomes [91], where the ARF-GEF facilitates the continuous recycling of PIN1 to the basal plasma membrane of plant cells [92], thereby underpinning polar auxin transport. Intriguingly, auxin itself appears to be a regulator of this constitutive recycling process, providing a feedback regulation mechanism for auxin efflux [93]. A recent article [94] additionally demonstrates that PIN2 is recycled through a endosomal compartment that is regulated by AtSNX1 and is distinct from the GNOM-regulated endosomal compartment. The basal polarity of PIN1 localization can be reversed by overexpressing the serine/threonine kinase protein PINOID [95], and there is some evidence that this may also affect PIN activity [71]. However, the substrate for PINOID phosphorylation and putative phosphatase enzymes remain to be identified.
The auxin-efflux carriers AtMDR1/PGP19 and AtMDR11/PGP1 interact with the immunophilin-like TWD1 (twisted dwarf 1) protein [96]. The phenotypes of the twd1 and atpgp1/atpgp19 mutants are very similar, suggesting that TWD1 regulates AtMDR1/PGP19 and AtMDR11/PGP1 function. TWD1 is unlikely to facilitate protein folding and/or trafficking, since the subcellular distribution of AtMDR11/PGP1 is comparable between wild-type and twd1-1 [96]. Instead, the plasma-membrane-localized TWD1 protein appears to directly regulate AtMDR1/PGP19 and AtMDR11/PGP1 auxin-transport function at the cell surface, a proposition which has been very recently confirmed [73]. The N-terminal domain of TWD1 has been shown to directly interact with C-terminal nucleotide-binding fold of AtMDR1/PGP19 and AtMDR11/PGP1 [96], suggesting that TWD1 regulates ATP binding and hydrolysis. Interestingly, the auxin-transport inhibitor NPA disrupts all TWD1–AtPGP interactions [96], highlighting the functional importance of this complex.
Knowledge about accessory proteins such as TWD1 and AXR4 is likely to be very important for heterologous expression studies. The failure to detect auxin-transport activity when expressing AtMDR1/PGP19 in yeast [48] could reflect a requirement for TWD1 to be co-expressed. Similarly, AUX1 took 6 days to accumulate in the plasma membrane when expressed in the Xenopus oocyte system [67], compared with other plant proteins, which normally take only 2 days, suggesting that AUX1 assembly was inefficient. AUX1 and AXR4 co-expression studies in Xenopus will reveal whether this is indeed the case.
CONCLUDING REMARKS
The past few years have seen significant strides forward in our attempts to describe the molecular basis and the regulation of the chemiosmotic polar transport of auxin (Figure 4). However, we do still have little mechanistic understanding of the various auxin transporters. We may be able to draw parallels from studies on related transport families. For example, detailed studies on ABC transporters, in terms of substrate recognition [49], transport mechanism [55] and structure [97], will surely guide future research on AtPGP proteins. Indeed, it remains possible that membrane-protein global-expression and crystallization projects will provide structural data for AUX/LAX and PIN relatives in advance of a full functional understanding of these proteins. Much intense research ahead also lies in the regulation of auxin-transport proteins. We need to identify any further auxiliary proteins which may either directly regulate auxin-transporter function or which may facilitate protein trafficking. The mechanism of these interactions also require elucidation, and potential parallels may emerge with studies of yeast amino acid permeases, which require the presence of an accessory protein to avoid inappropriate interactions during translation and insertion into the ER membrane [98]. The precise role of ABC transporters in auxin efflux (and influx?) remains to be established. That AtPGPs function as auxin transporters seems assured, but are their properties modified through interactions with PINs [66] (and perhaps with AUX/LAX proteins)? Examples of mammalian ABC transporters involved in complexes which are required for both protein targeting and for protein function have been described [99], and co-expression studies may be able to reveal whether the same is true for AtPGP proteins. Clearly an additional impetus is now to find structure–function relationships for the auxin-transport proteins. Only one significant allelic series exists for an auxin-transport protein, namely for AUX1 [28], and very few mutant PINs and auxin-transporting ABC proteins have been described [64,69]. The existence of convincing heterologous-expression-system transport data now permits expansion into this area, with the promise of providing a kinetic description of transport. Understanding the kinetics of the auxin transporters will make a great contribution to attempts to mathematically model auxin transport and distribution [100], with concomitant insights into plant regulation.
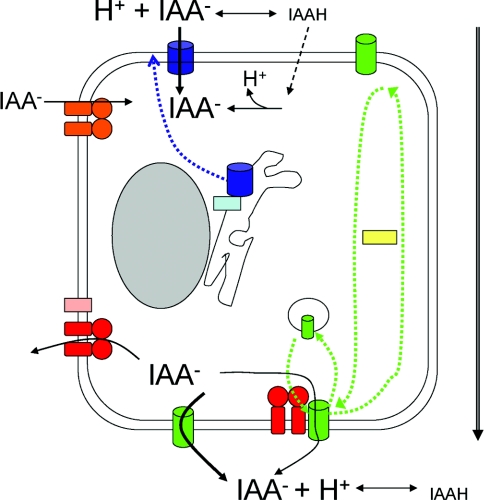
Figure 4. Function and regulation of auxin-transport proteins.
Auxin enters the cell primarily through carrier-mediated influx via AUX1 (blue cylinder) as the anionic form, with a small diffusional contribution of the uncharged species (broken line). Auxin efflux is mediated via PIN proteins (green cylinders) with some contribution from AtPGP1/19 (red), possibly via direct protein–protein interactions. AUX1 regulation by the ER protein AXR4 (light-blue box) is shown as affecting the trafficking of AUX1 to the plasma membrane (dotted blue arrow). PIN polarity controlled by PINOID (yellow box) and an unconfirmed phosphatase are depicted by green arrows, whereas the endosomal cycling of PIN1 and PIN2 is shown schematically by the intracellular vesicle. AtPGP1/19 interactions with TWD (pink box) and the putative import of IAA through AtPGP4 (orange) are also depicted. The overall direction of polar auxin transport is shown by the large arrow to the right-hand side.
Acknowledgments
I.D.K. and M.J.B. thank the BBSRC (Biotechnology and Biological Sciences Research Council) for project grant support (grant no. BB/C514958/1). We declare no competing financial interests.
References
- 1.Pennazio S. The discovery of the chemical nature of the plant hormone auxin. Riv. Biol. 2002;95:289–308. [PubMed] [Google Scholar]
- 2.Perrot-Rechenmann C., Napier R. M. Auxins. Vitam. Horm. 2005;72:203–233. doi: 10.1016/S0083-6729(04)72006-3. [DOI] [PubMed] [Google Scholar]
- 3.Dharmasiri N., Dharmasiri S., Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 4.Kepinski S., Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 5.Woodward A. W., Bartel B. A receptor for auxin. Plant Cell. 2005;17:2425–2429. doi: 10.1105/tpc.105.036236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruegger M., Dewey E., Gray W. M., Hobbie L., Turner J., Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ljung K., Bhalerao R. P., Sandberg G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 2001;28:465–474. doi: 10.1046/j.1365-313x.2001.01173.x. [DOI] [PubMed] [Google Scholar]
- 8.Friml J., Palme K. Polar auxin transport – old questions and new concepts? Plant Mol. Biol. 2002;49:273–284. [PubMed] [Google Scholar]
- 9.Ljung K., Hull A. K., Celenza J., Yamada M., Estelle M., Normanly J., Sandberg G. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell. 2005;17:1090–1104. doi: 10.1105/tpc.104.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benkova E., Michniewicz M., Sauer M., Teichmann T., Seifertova D., Jurgens G., Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 11.Swarup R., Bennett M. Auxin transport: the fountain of life in plants? Dev. Cell. 2003;5:824–826. doi: 10.1016/s1534-5807(03)00370-8. [DOI] [PubMed] [Google Scholar]
- 12.Swarup R., Kramer E. M., Perry P., Knox K., Leyser H. M., Haseloff J., Beemster G. T., Bhalerao R., Bennett M. J. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 2005;7:1057–1065. doi: 10.1038/ncb1316. [DOI] [PubMed] [Google Scholar]
- 13.Estelle M. Plant tropisms: the ins and outs of auxin. Curr. Biol. 1996;6:1589–1591. doi: 10.1016/s0960-9822(02)70780-x. [DOI] [PubMed] [Google Scholar]
- 14.Kramer E. M., Bennett M. J. Auxin transport: a field in flux. Trends Plant Sci. 2006;11:382–386. doi: 10.1016/j.tplants.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Rubery P. H., Sheldrake A. R. Carrier-mediated auxin transport. Planta. 1974;118:101–121. doi: 10.1007/BF00388387. [DOI] [PubMed] [Google Scholar]
- 16.Blakeslee J. J., Peer W. A., Murphy A. S. Auxin transport. Curr. Opin. Plant Biol. 2005;8:494–500. doi: 10.1016/j.pbi.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Parry G., Delbarre A., Marchant A., Swarup R., Napier R., Perrot-Rechenmann C., Bennett M. J. Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1. Plant J. 2001;25:399–406. doi: 10.1046/j.1365-313x.2001.00970.x. [DOI] [PubMed] [Google Scholar]
- 18.Marchant A., Kargul J., May S. T., Muller P., Delbarre A., Perrot-Rechenmann C., Bennett M. J. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999;18:2066–2073. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galweiler L., Guan C., Muller A., Wisman E., Mendgen K., Yephremov A., Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 20.Bennett M. J., Marchant A., Green H. G., May S. T., Ward S. P., Millner P. A., Walker A. R., Schulz B., Feldmann K. A. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- 21.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 22.Schnabel E. L., Frugoli J. The PIN and LAX families of auxin transport genes in Medicago truncatula. Mol. Genet. Genomics. 2004;272:420–432. doi: 10.1007/s00438-004-1057-x. [DOI] [PubMed] [Google Scholar]
- 23.Goff S. A., Ricke D., Lan T. H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 24.Yu J., Hu S., Wang J., Wong G. K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 25.Swarup R., Friml J., Marchant A., Ljung K., Sandberg G., Palme K., Bennett M. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 2001;15:2648–2653. doi: 10.1101/gad.210501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young G. B., Jack D. L., Smith D. W., Saier M. H., Jr The amino acid/auxin:proton symport permease family. Biochim. Biophys. Acta. 1999;1415:306–322. doi: 10.1016/s0005-2736(98)00196-5. [DOI] [PubMed] [Google Scholar]
- 27.Prusty R., Grisafi P., Fink G. R. The plant hormone indoleacetic acid induces invasive growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4153–4157. doi: 10.1073/pnas.0400659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swarup R., Kargul J., Marchant A., Zadik D., Rahman A., Mills R., Yemm A., May S., Williams L., Millner P., et al. Structure–function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell. 2004;16:3069–3083. doi: 10.1105/tpc.104.024737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moller S., Croning M. D., Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 30.Hong R., Sansom M. S. Computer prediction of transporter topology and structure. In: Baldwin S. A., editor. Membrane Transport: A Practical Approach. Oxford: Oxford University Press; 2000. pp. 209–228. [Google Scholar]
- 31.Chang H. C., Bush D. R. Topology of NAT2, a prototypical example of a new family of amino acid transporters. J. Biol. Chem. 1997;272:30552–30557. doi: 10.1074/jbc.272.48.30552. [DOI] [PubMed] [Google Scholar]
- 32.Donnelly D. The arrangement of the transmembrane helices in the secretin receptor family of G-protein-coupled receptors. FEBS Lett. 1997;409:431–436. doi: 10.1016/s0014-5793(97)00546-2. [DOI] [PubMed] [Google Scholar]
- 33.Zheng L., Kostrewa D., Berneche S., Winkler F. K., Li X. D. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luschnig C., Gaxiola R. A., Grisafi P., Fink G. R. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Utsuno K., Shikanai T., Yamada Y., Hashimoto T. Agr, an Agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant Cell Physiol. 1998;39:1111–1118. doi: 10.1093/oxfordjournals.pcp.a029310. [DOI] [PubMed] [Google Scholar]
- 36.Chen R., Hilson P., Sedbrook J., Rosen E., Caspar T., Masson P. H. The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15112–15117. doi: 10.1073/pnas.95.25.15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller A., Guan C., Galweiler L., Tanzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998;17:6903–6911. doi: 10.1093/emboj/17.23.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paponov I. A., Teale W. D., Trebar M., Blilou I., Palme K. The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci. 2005;10:170–177. doi: 10.1016/j.tplants.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Donnelly D., Overington J. P., Ruffle S. V., Nugent J. H., Blundell T. L. Modeling α-helical transmembrane domains: the calculation and use of substitution tables for lipid-facing residues. Protein Sci. 1993;2:55–70. doi: 10.1002/pro.5560020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labarre C., Guzzo J., Cavin J. F., Divies C. Cloning and characterization of the genes encoding the malolactic enzyme and the malate permease of Leuconostoc oenos. Appl. Environ. Microbiol. 1996;62:1274–1282. doi: 10.1128/aem.62.4.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoenke S., Schmid M., Dimroth P. Sequence of a gene cluster from Klebsiella pneumoniae encoding malonate decarboxylase and expression of the enzyme in Escherichia coli. Eur. J. Biochem. 1997;246:530–538. doi: 10.1111/j.1432-1033.1997.00530.x. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Fernandez R., Emyr Davies T. G., Coleman J. O. D., Rea P. A. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- 43.Decottignies A., Goffeau A. Complete inventory of the yeast ABC proteins. Nat. Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- 44.Dean M., Rzhetsky A., Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 45.Geisler M., Blakeslee J. J., Bouchard R., Lee O. R., Vincenzetti V., Bandyopadhyay A., Titapiwatanakun B., Peer W. A., Bailly A., Richards E. L., et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005;44:179–194. doi: 10.1111/j.1365-313X.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- 46.Gadsby D. C., Vergani P., Csanady L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nichols C. G. KATP channels as molecular sensors of cellular metabolism. Nature. 2006;440:470–476. doi: 10.1038/nature04711. [DOI] [PubMed] [Google Scholar]
- 48.Noh B., Murphy A. S., Spalding E. P. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell. 2001;13:2441–2454. doi: 10.1105/tpc.010350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Veen H. W., Callaghan R. Substrate-binding sites in ABC transporters. In: Holland I. B., Cole S. P. C., Kuchler K., Higgins C. F., editors. ABC Proteins: From Bacteria to Man. New York: Academic Press; 2003. pp. 81–105. [Google Scholar]
- 50.Sidler M., Hassa P., Hasan S., Ringli C., Dudler R. Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell. 1998;10:1623–1636. doi: 10.1105/tpc.10.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tusnady G. E., Sarkadi B., Simon I., Varadi A. Membrane topology of human ABC proteins. FEBS Lett. 2006;580:1017–1022. doi: 10.1016/j.febslet.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 52.Linton K. J., Rosenberg M. F., Kerr I. D., Higgins C. F. Structure of ABC transporters. In: Holland I. B., Cole S. P. C., Kuchler K., Higgins C. F., editors. ABC Proteins: From Bacteria to Man. New York: Academic Press; 2003. pp. 65–80. [Google Scholar]
- 53.Kerr I. D. Structure and association of ATP binding cassette transporter nucleotide-binding domains. Biochem. Biophys. Acta. 2002;1561:47–64. doi: 10.1016/s0304-4157(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 54.Schmitt L., Tampe R. Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 2002;12:754–760. doi: 10.1016/s0959-440x(02)00399-8. [DOI] [PubMed] [Google Scholar]
- 55.Callaghan R., Ford R. C., Kerr I. D. The translocation mechanism of P-glycoprotein. FEBS Lett. 2006;580:1056–1063. doi: 10.1016/j.febslet.2005.11.083. [DOI] [PubMed] [Google Scholar]
- 56. Reference deleted.
- 57.Lin R., Wang H. Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol. 2005;138:949–964. doi: 10.1104/pp.105.061572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Multani D. S., Briggs S. P., Chamberlin M. A., Blakeslee J. J., Murphy A. S., Johal G. S. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science. 2003;302:81–84. doi: 10.1126/science.1086072. [DOI] [PubMed] [Google Scholar]
- 59.Santelia D., Vincenzetti V., Azzarello E., Bovet L., Fukao Y., Duchtig P., Mancuso S., Martinoia E., Geisler M. MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett. 2005;579:5399–5406. doi: 10.1016/j.febslet.2005.08.061. [DOI] [PubMed] [Google Scholar]
- 60.Terasaka K., Blakeslee J. J., Titapiwatanakun B., Peer W. A., Bandyopadhyay A., Makam S. N., Lee O. R., Richards E. L., Murphy A. S., Sato F., Yazaki K. PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell. 2005;17:2922–2939. doi: 10.1105/tpc.105.035816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gaedeke N., Klein M., Kolukisaoglu U., Forestier C., Muller A., Ansorge M., Becker D., Mamnun Y., Kuchler K., Schulz B., et al. The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J. 2001;20:1875–1887. doi: 10.1093/emboj/20.8.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klein M., Perfus-Barbeoch L., Frelet A., Gaedeke N., Reinhardt D., Mueller-Roeber B., Martinoia E., Forestier C. The plant multidrug resistance ABC transporter AtMRP5 is involved in guard cell hormonal signalling and water use. Plant J. 2003;33:119–129. doi: 10.1046/j.1365-313x.2003.016012.x. [DOI] [PubMed] [Google Scholar]
- 63.Zolman B. K., Silva I. D., Bartel B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 2001;127:1266–1278. [PMC free article] [PubMed] [Google Scholar]
- 64.Ito H., Gray W. M. A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 2006;142:63–74. doi: 10.1104/pp.106.084533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mikhailov M. V., Campbell J. D., de Wet H., Shimomura K., Zadek B., Collins R. F., Sansom M. S., Ford R. C., Ashcroft F. M. 3-D structural and functional characterization of the purified KATP channel complex Kir6.2-SUR1. EMBO J. 2005;24:4166–4175. doi: 10.1038/sj.emboj.7600877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noh B., Bandyopadhyay A., Peer W. A., Spalding E. P., Murphy A. S. Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature. 2003;423:999–1002. doi: 10.1038/nature01716. [DOI] [PubMed] [Google Scholar]
- 67.Yang Y., Hammes U. Z., Taylor C. G., Schachtman D. P., Nielsen E. High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 2006;16:1123–1127. doi: 10.1016/j.cub.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 68.Imhoff V., Muller P., Guern J., Delbarre A. Inhibitors of the carrier-mediated influx of auxin in suspension-cultured tobacco cells. Planta. 2000;210:580–588. doi: 10.1007/s004250050047. [DOI] [PubMed] [Google Scholar]
- 69.Petrasek J., Mravec J., Bouchard R., Blakeslee J. J., Abas M., Seifertova D., Wisniewska J., Tadele Z., Kubes M., Covanova M., et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312:914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- 70.Petrasek J., Cerna A., Schwarzerova K., Elckner M., Morris D. A., Zazimalova E. Do phytotropins inhibit auxin efflux by impairing vesicle traffic? Plant Physiol. 2003;131:254–263. doi: 10.1104/pp.012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee S. H., Cho H. T. PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell. 2006;18:1604–1616. doi: 10.1105/tpc.105.035972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Decottignies A., Grant A. M., Nichols J. W., de Wet H., McIntosh D. B., Goffeau A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 1998;273:12612–12622. doi: 10.1074/jbc.273.20.12612. [DOI] [PubMed] [Google Scholar]
- 73.Bouchard R., Bailly A., Blakeslee J. J., Vincenzetti V., Paponov I., Palme K., Mancuso S., Murphy A. S., Schulz B., Geisler M. Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J. Biol. Chem. 2006;281:30603–30612. doi: 10.1074/jbc.M604604200. [DOI] [PubMed] [Google Scholar]
- 74.Loo T. W., Clarke D. M. Superfolding of the partially unfolded core-glycosylated intermediate of human P-glycoprotein into the mature enzyme is promoted by substrate-induced transmembrane domain interactions. J. Biol. Chem. 1998;273:14671–14674. doi: 10.1074/jbc.273.24.14671. [DOI] [PubMed] [Google Scholar]
- 75.Stenham D. R., Campbell J. D., Sansom M. S. P., Higgins C. F., Kerr I. D., Linton K. J. An atomic detail model for the human ATP binding cassette transporter, P-glycoprotein, derived from disulphide cross-linking and homology modelling. FASEB J. 2003;15:2287–2289. doi: 10.1096/fj.03-0107fje. [DOI] [PubMed] [Google Scholar]
- 76.Martin C., Berridge G., Higgins C. F., Mistry P., Charlton P., Callaghan R. Communication between multiple drug binding sites on P-glycoprotein. Mol. Pharmacol. 2000;58:624–632. doi: 10.1124/mol.58.3.624. [DOI] [PubMed] [Google Scholar]
- 77.Shapiro A. B., Ling V. Positively cooperative sites for drug transport by P-glycoprotein with distinct drug specificities. Eur. J. Biochem. 1997;250:130–137. doi: 10.1111/j.1432-1033.1997.00130.x. [DOI] [PubMed] [Google Scholar]
- 78.Lankat Buttgereit B., Tampe R. The transporter associated with antigen processing TAP: structure and function. FEBS Lett. 1999;464:108–112. doi: 10.1016/s0014-5793(99)01676-2. [DOI] [PubMed] [Google Scholar]
- 79.Shitan N., Bazin I., Dan K., Obata K., Kigawa K., Ueda K., Sato F., Forestier C., Yazaki K. Involvement of CjMDR1, a plant multidrug-resistance-type ATP-binding cassette protein, in alkaloid transport in Coptis japonica. Proc. Natl. Acad. Sci. U.S.A. 2003;100:751–756. doi: 10.1073/pnas.0134257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Urbatsch I. L., Sankaran B., Weber J., Senior A. E. P-glycoprotein is stably inhibited by vanadate-induced trapping of nucleotide at a single catalytic site. J. Biol. Chem. 1995;270:19383–19390. doi: 10.1074/jbc.270.33.19383. [DOI] [PubMed] [Google Scholar]
- 81.Urbatsch I. L., Sankaran B., Bhagat S., Senior A. E. Both P-glycoprotein nucleotide-binding sites are catalytically active. J. Biol. Chem. 1995;270:26956–26961. doi: 10.1074/jbc.270.45.26956. [DOI] [PubMed] [Google Scholar]
- 82.Urbatsch I. L., Julien M., Carrier I., Rousseau M.-E., Cayrol R., Gros P. Mutational analysis of conserved carboxylate residuesin the nucleotide binding sites of P-glycoprotein. Biochemistry. 2000;39:14138–14149. doi: 10.1021/bi001128w. [DOI] [PubMed] [Google Scholar]
- 83.Martin C., Berridge G., Mistry P., Higgins C. F., Charlton P., Callaghan R. Drug binding sites on P-glycoprotein are altered by ATP binding prior to nucleotide hydrolysis. Biochemistry. 2000;39:11901–11906. doi: 10.1021/bi000559b. [DOI] [PubMed] [Google Scholar]
- 84.Clark R., Kerr I. D., Callaghan R. Multiple drug binding sites on the ABCG2 transporter. Br. J. Pharmacol. 2006;149:506–515. doi: 10.1038/sj.bjp.0706904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dharmasiri S., Swarup R., Mockaitis K., Dharmasiri N., Singh S. K., Kowalchyk M., Marchant A., Mills S., Sandberg G., Bennett M. J., Estelle M. AXR4 is required for localization of the auxin influx facilitator AUX1. Science. 2006;312:1218–1220. doi: 10.1126/science.1122847. [DOI] [PubMed] [Google Scholar]
- 86.Hobbie L. Auxin and cell polarity: the emergence of AXR4. Trends Plant Sci. 2006;11:517–518. doi: 10.1016/j.tplants.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 87.Molendijk A. J., Ruperti B., Palme K. Small GTPases in vesicle trafficking. Curr. Opin. Plant Biol. 2004;7:694–700. doi: 10.1016/j.pbi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 88.Shevell D. E., Leu W. M., Gillmor C. S., Xia G., Feldmann K. A., Chua N. H. EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell. 1994;77:1051–1062. doi: 10.1016/0092-8674(94)90444-8. [DOI] [PubMed] [Google Scholar]
- 89.Busch M., Mayer U., Jurgens G. Molecular analysis of the Arabidopsis pattern formation of gene GNOM: gene structure and intragenic complementation. Mol. Gen. Genet. 1996;250:681–691. doi: 10.1007/BF02172979. [DOI] [PubMed] [Google Scholar]
- 90.Donaldson J. G., Jackson C. L. Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 2000;12:475–482. doi: 10.1016/s0955-0674(00)00119-8. [DOI] [PubMed] [Google Scholar]
- 91.Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jurgens G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112:219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 92.Geldner N., Friml J., Stierhof Y. D., Jurgens G., Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- 93.Paciorek T., Zazimalova E., Ruthardt N., Petrasek J., Stierhof Y. D., Kleine-Vehn J., Morris D. A., Emans N., Jurgens G., Geldner N., Friml J. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature. 2005;435:1251–1256. doi: 10.1038/nature03633. [DOI] [PubMed] [Google Scholar]
- 94.Jaillais Y., Fobis-Loisy I., Miege C., Rollin C., Gaude T. AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature. 2006;443:104–107. doi: 10.1038/nature05046. [DOI] [PubMed] [Google Scholar]
- 95.Friml J., Yang X., Michniewicz M., Weijers D., Quint A., Tietz O., Benjamins R., Ouwerkerk P. B., Ljung K., Sandberg G., et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- 96.Geisler M., Kolukisaoglu H. U., Bouchard R., Billion K., Berger J., Saal B., Frangne N., Koncz-Kalman Z., Koncz C., Dudler R., Blakeslee J. J., et al. TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell. 2003;14:4238–4249. doi: 10.1091/mbc.E02-10-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dawson R. J., Locher K. P. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 98.Kota J., Ljungdahl P. O. Specialized membrane-localized chaperones prevent aggregation of polytopic proteins in the ER. J. Cell Biol. 2005;168:79–88. doi: 10.1083/jcb.200408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zerangue N., Schwappach B., Jan Y. N., Jan L. Y. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane KATP channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 100.Kramer E. M. PIN and AUX/LAX proteins: their role in auxin accumulation. Trends Plant Sci. 2004;9:578–582. doi: 10.1016/j.tplants.2004.10.010. [DOI] [PubMed] [Google Scholar]