Abstract
Centaurins are a family of proteins that contain GTPase-activating protein domains, with the γ family members containing in addition a GTPase-like domain. Centaurins reside mainly in the nucleus and are known to activate phosphoinositide 3-kinase, a key regulator of cell proliferation, motility and vesicular trafficking. In the present study, using X-ray structural analysis, enzymatic assays and nucleotide-binding studies, we show that, for CENTG1 (centaurin γ-1) the GTPase-like domain has broader trinucleotide specificity. Alterations within the G4 motif of CENTG1 from the highly conserved NKXD found in typical GTPases to TQDR result in the loss of specificity, a lower affinity for the nucleotides and higher turnover rates. These results indicate that the centaurins could be more accurately classified as NTPases and point to alternative mechanisms of cell signalling control.
Keywords: centaurin γ-1 (CENTG1), GTPase-activating domain, NTPase, phosphoinositide 3-kinase (PI3K), phosphoinositide 3-kinase enhancer (PIKE)
Abbreviations: ATP[S], adenosine 5′-[γ-thio]triphosphate; CENTG, centaurin γ; DTT, dithiothreitol; GAP, GTPase-activating protein; GEF, guanine-nucleotide-exchange factor; gi, GenInfo Identifier; GLD, GTPase-like domain; GTP[S], guanosine 5′-[γ-thio]triphosphate; ITC, isothermal calorimetry; Ni-NTA, Ni2+-nitrilotriacetate; PEG1000, poly(ethylene glycol) 1000; PH, pleckstrin homology; PI3K, phosphoinositide 3-kinase; (r)PIKE, (rat) PI3K enhancer; rmsd, root mean square deviation; TEV, tobacco etch virus
INTRODUCTION
Cell signalling involves the detection, amplification and regulation of a specific signal that frequently starts when a ligand binds to its cell-surface receptor. In the case of the family of small GTPases, activation of the signal pathway involves a switch of the GTPase from an inactive GDP-bound form to the active GTP-bound state [1–3]. The ability of the GTPases to function as molecular switches is regulated through differential action of GEFs (guanine-nucleotide-exchange factors) and GAPs (GTPase-activating proteins). GEFs convert the GTPases into the active state by replacing bound GDP for GTP, whereas GAPs reverse this process and down-regulate the signalling pathway by enhancing the GTPase activity, thus converting the enzyme into the inactive GDP-bound state. Differences in structure between the GDP- and GTP-bound states of the GTPases are the basis for this signal transmission. The main changes centre on two loop regions designated Switch I and Switch II [4–8]. There are five subfamilies based on sequence alignment (Ras, Rho, Rab, Arf/Sar1 and Ran) that comprise the small GTPase family [9].
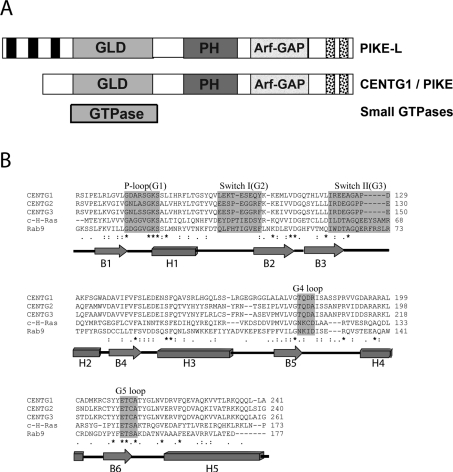
All of the centaurin proteins, like many proteins involved in signalling, are composed of multiple domains. The domains that the centaurin family members have in common include an Arf-GAP domain, ankyrin repeats and PH (pleckstrin homology). Both the cytohesin and centaurin PH domains bind the lipid phosphatidylinositol 3,4,5-trisphosphate [10,11], thus localizing them to the membrane surface where their target protein is located. The centaurin γ subfamily stands out from the rest of the centaurins for having an unusual domain organization with a GLD (GTPase-like domain) on the N-terminus (Figure 1A). On the basis of sequence identity, this GTPase domain is actually more closely related to the Rab and Ras subfamilies than to the Arf family of small GTPases. The human CENTG (centaurin γ) subfamily is represented by three isoforms, CENTG1 (centaurin γ-1), CENTG2 (centaurin γ-2) and CENTG3 (centaurin γ-3), which are highly similar in primary structure (Figure 1B). Owing to the sequence similarity of the GLD to the classical GTPase family and the presence of an Arf-GAP domain, it has been assumed that the CENTG GLDs are functionally GTPases.
Figure 1. Domain and sequence comparison of CENTG1.
(A) Domain architecture of the centaurins and small GTPases. PIKE isoform-A or CENTG1 is shown in comparison with long isoform PIKE-L and small GTPases. The proline-rich domain present only in PIKE-L is shown in black rectangles at the N-terminus. The GLDs of CENTG1 and PIKE-L, and GTPase domain of a small GTPase are shown in light grey. The PH and Arf-GAP domains are indicated. Two ankyrin repeats in the C-terminus are shown as black dot-filled boxes. (B) Sequence alignment of human CENTG with small GTPases. ClustalW-based sequence alignment of CENTG1 (gi:6176569), CENTG2 (gi:51338837) and CENTG3 (gi:16799069) with the small GTPases c-H-Ras (gi:231061) and Rab9A (gi:55670684), with editing to reflect the structural superimposition. Identical amino acids are indicated by an asterisk (*). Highly conserved small GTPase functional loops, G1–G5, are highlighted in grey. The secondary-structure depiction beneath the sequences is according to the CENTG1 GLD structure determined here (PDB code 2BMJ). α-Helices are represented as rectangles and β-sheets as arrows.
The rPIKE [rat PI3K (phosphoinositide 3-kinase) enhancer] proteins are recognized to be the closest homologues to the CENTG proteins according to gene structure and sequence (Figure 1). rPIKE is a brain-specific nuclear GTPase protein, which binds PI3K and stimulates its lipid-binding activity [12]. Three different alternatively spliced forms of PIKE have been reported: PIKE-L (long isoform) [gi (GenInfo Identifier): 25989575], PIKE-S (short isoform) and the recently identified isoform called PIKE-A (gi:1531538). The PIKE-A isoform is the product of a different initiation transcript of PIKE-L and is identical with the human CENTG1. CENTG1 has been called GGAP2 previously, whereas CENTG2 has been called GGAP1 previously. We refer to these proteins as CENTG1 and CENTG2 in the present paper.
CENTG1 was shown to interact directly with activated PKB (protein kinase B)/Akt in a GTP-dependent manner and stimulate Akt kinase activity [13]. CENTG1 is co-amplified with CDK4 (cyclin-dependent kinase 4) in a variety of human cancers [14,15]. Recent studies have shown that CENTG1 is a physiological regulator of apoptosis [16] and an oncogenic effector of cell invasion through its activation of Akt function [13].
The signalling pathways mediated by the Ras superfamily in the cell cytoplasm have been characterized extensively. However, the mode of action of signalling GTPases in the nucleus remains largely obscure. In the present paper, we show the crystal structure of the N-terminal GLD of CENTG1 in the absence of bound nucleotide. Significant structural differences exist between the CENTG1 GLD and the archetypal small GTPases, such as H-ras, that called into question the nucleotide-specificity for this GLD. To verify its activity, we performed enzymatic assays that indicated that the domain is able to catalyse the hydrolysis of a selection of triphosphate nucleotides, thereby classifying the CENTG1 GLD as an NTPase. No other human GTPase-like domain showing NTPase activity has been characterized before. The present study provides a beginning for the understanding of the structure and function of this family of nuclear NTPases that actively participates in the promotion of cancer cell invasion and prevention of apoptosis.
EXPERIMENTAL
Cloning
A sequence containing the GLD of CENTG1 (gi:7661962) was amplified by PCR from DNA in the Mammalian Gene Collection (I.M.A.G.E. Consortium Clone ID 5218187; gi:20380827) and subcloned into an in-house vector carrying ampicillin resistance, pLIC-SGC, using ligation-independent cloning. The resulting plasmid expresses residues 66–241 of the GLD with an N-terminal hexahistidine tag and TEV (tobacco etch virus) protease tag cleavage site (extension MHHHHHHSSGVDLGTENLYFQ*SM-). After digestion with TEV protease, the protein retains an additional serine and a methionine residue on the N-terminus. The point mutants of CENTG1 were created in a two-step overlapping PCR process and were subcloned into pLIC-SGC in an identical manner. The sequences of all constructs were confirmed by DNA sequencing.
Expression and purification
Plasmids encoding the CENTG1 GLD or the mutants were transformed into Escherichia coli BL21(DE3) competent cells and the transformants were used to inoculate 10 ml of Luria–Bertani medium with 100 μg/ml ampicillin which was incubated overnight at 37 °C. This culture was used to inoculate 1 litre of Terrific broth with 100 μg/ml ampicillin and grown at 37 °C until a D600 of 0.6 was reached. The protein was induced with 1 mM IPTG (isopropyl β-D-thiogalactoside) for 12 h at 18 °C. The cells were harvested by centrifugation at 9000 g for 20 min using a Beckman Coulter JLA-8.100 rotor, resuspended in lysis buffer (20 mM Tris/HCl, pH 8.0, 200 mM NaCl, 5% glycerol and 10 mM imidazole) and frozen at −80 °C until further use.
The frozen cell pellet was thawed in the presence of one EDTA-free Complete™ protease inhibitor tablet (Roche). The cells were lysed using an Emulsiflex C5 high pressure homogenizer (Avestin), poly(ethyleneimine) was added to a final concentration of 0.15%, and the insoluble debris was removed by centrifugation for 45 min at 15000 rev./min using a Beckman Coulter JA-17 rotor. The CENTG1 GLD was extracted from the clarified supernatant by affinity-tag purification using Ni-NTA (Ni2+-nitrilotriacetate) resin (Qiagen): the supernatant was bound to Ni-NTA and washed with 30 column vol. of lysis buffer and 5 column vol. of wash buffer (20 mM Tris/HCl, pH 8.0, 200 mM NaCl, 5% glycerol and 20 mM imidazole). The protein was eluted from the resin with 5 column vol. of elution buffer (20 mM Tris/HCl, pH 8.0, 200 mM NaCl, 5% glycerol and 150 mM imidazole). The eluted protein was purified further by gel-filtration chromatography (using an S75 16/60 column) in either 50 mM Tris/HCl, pH 8.0, and 150 mM NaCl (for the C2 space group) or 50 mM Tris/HCl, pH 8.0, 150 mM NaCl, 100 μM GDP, 10 mM NaF, 20 μM AlF3 and 5 mM DTT (dithiothreitol) (for the P212121 space group). Fractions containing the CENTG1 GLD were identified using SDS/PAGE (4–20% gradient gels), pooled and concentrated to 10 mg/ml using a 10 kDa cut-off concentrator. The protein was incubated overnight at 4 °C with TEV protease and purified from the tag by passing over Ni-NTA. The purified CENTG1 GLD was concentrated to 30 mg/ml. The identities of the CENTG1 GLD proteins were confirmed by MS under denaturing conditions using an Agilent LC-MS system with a reverse-phase column (wild-type: expected molecular mass 19581.1 Da, observed molecular mass 19581.4 Da; T101A: expected molecular mass 19551.0 Da, observed molecular mass 19550.8 Da; T101P: expected molecular mass 19577.1 Da, observed molecular mass 19577.0 Da).
Crystallization, data collection and structure solution
Crystals of CENTG1 GLD were obtained from 150 nl sitting drops containing a 1:1 ratio of protein to reservoir solution at 20 °C. The reservoir solution consisted of either 50 mM Tris/HCl, pH 7.6, 150 mM NaCl, 0.1 M SPG (sucrose/phosphate/glutamate), 30% PEG1000 [poly(ethylene glycol) 1000] and 0.5% DMSO (C2 space group) or 0.1 M propionate/cacodylate/Bis-Tris propane, pH 6.0, and 30% PEG1000 (P212121 space group). Crystals were mounted in nylon loops and cryo-cooled by transfer to a 100 K nitrogen stream. Data were collected on a Rigaku/MSC FR-E rotating anode generator equipped with an R-AXIS HTC image plate.
The data were processed using MOSFLM [17] and the CCP4 suite [18]. A total of 5% of the reflections were excluded for calculation of Rfree. A molecular replacement solution was found for the C2 space group using PHASER [19] and an ensemble of search models with primary sequence identity with CENTG1 between 25 and 27%. There was one molecule in the asymmetric unit. The model was rebuilt using O [20] and refinement was performed using CNS [21] and finally Refmac5 [22]. The P212121 space group was solved by molecular replacement using the refined structure from the C2 space group as a search model. There was one molecule in the asymmetric unit. Data processing and refinement statistics can be found in Table 1.
Table 1. Data collection and refinement.
Numbers in parentheses for data collection refer to the highest resolution shell.
| Parameter | Value | |
|---|---|---|
| Data collection | ||
| Space group | C2 | P212121 |
| Unit cell [a, b, c (Å), β (°)] | 62.2, 31.1, 81.9, 102.2 | 36.3, 62.1, 67.5, 90.0 |
| Resolution range (Å) | 30.63-2.10 (2.21–2.10) | 20.72-1.50 (1.58–1.50) |
| Number of unique observations | 8653 (1180) | 24972 (3424) |
| Number of total observations | 43169 (5102) | 217987 (14791) |
| Completeness (%) | 95.1 (89.9) | 99.3 (95.6) |
| Multiplicity | 5.0 (4.3) | 8.7 (4.3) |
| Rmerge (%) | 6.8 (22.1) | 6.0 (33.7) |
| 〈I/σ(I)〉 | 16.9 (5.6) | 24.8 (2.7) |
| Refinement | ||
| R factor (%) | 16.3 | 15.0 |
| Rfree (%) | 22.5 | 21.4 |
| Rmsd bond length (Å) [angle (°)] | 0.014 (1.53) | 0.011 (1.47) |
| PDB code | 2BMJ | 2IWR |
Relative activity of gel-filtration fractions
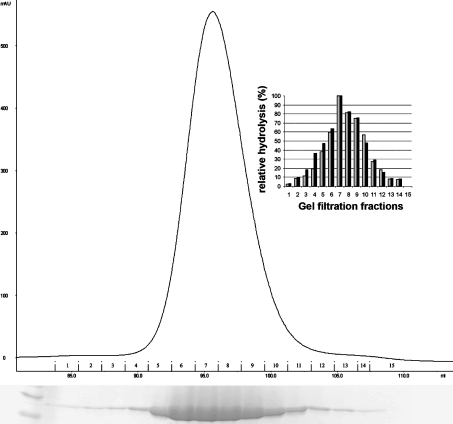
The purified protein was gel-filtered and the eluate fractions were 99% pure when analysed on SDS denaturing gels. The protein from each fraction was analysed for GTP and NTP hydrolysis using a Malachite Green phosphate assay kit (BioAssay Systems), according to the manufacturer's instructions. The release of phosphate was quantified using a standard curve, and the relative GTPase and NTPase activity of each fraction was determined and plotted as shown in Figure 2. Since the altered nucleotide specificity was unusual for GTPases, this procedure was carried out in order to ensure that there was no contaminating enzyme contributing to the NTPase activity.
Figure 2. GTPase and NTPase activity of the gel-filtration fractions of the CENTG1 GLD.
Chromatogram of the Superdex 75 gel-filtration run with the Coomassie Blue-stained SDS/PAGE gel corresponding to the fractions displayed beneath. The extreme left of the gel shows the molecular-mass markers. The measured activity was plotted for every fraction. The black and grey bars in the inset histogram represent the percentage of GTPase and NTPase activity of each fraction respectively.
GTPase and NTPase activity assay
Purified CENTG1 GLD (100 nM) was incubated with 12 μM–5 mM nucleotide triphosphates in reaction buffer (20 mM Hepes, pH 7.0, 150 mM NaCl and 5 mM MgCl2) at 30 °C in an 80 μl reaction volume. The assays performed with 1 and 2 mM MgCl2 did not show any difference in activity. The reactions were repeated at pH 6.0 and 8.0. The experiment was performed according to the manufacturer's instructions. The product formation was determined by recording the absorption at 650 nm using a SPECTRAmax spectrophotometer (Molecular Devices) 1 min after the initiation of the reaction. The experiment was repeated with different substrate concentrations. Blanks containing corresponding nucleotide concentrations in the reaction buffer with Malachite Green solution were subtracted from each recording. To determine values for Vmax and Km, the data were fitted to the Michaelis–Menten equation using non-linear regression (Kaleidograph software). All nucleotides and analogues were purchased from Sigma.
Non-covalent MS
Non-covalent MS was performed on a Q-Tof Micro (Waters) instrument equipped with a Nanomate spray chip (Advion) nano-electrospray ionization source. Samples were buffer-exchanged into aqueous ammonium acetate buffer (10–25 mM, pH 7) using micro-biospin chromatography columns (Bio-Rad). Pressure settings on the instrument were approx. 60 Pa (source, Speedivalve) with sample and extractor cone voltages chosen so that non-covalent complexes were preserved intact [23,24].
ITC (isothermal calorimetry)
ITC measurements were carried out at 20 °C using a VP-ITC MicroCalorimeter (MicroCal). CENTG1 and the nucleotides were in a solution of 20 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM MgCl2 and 5 mM DTT, which was degassed in a ThermoVac apparatus (MicroCal). ITC experiments were performed by stepwise titration of the non-hydrolysable NTP analogue GTP[S] (guanosine 5′-[γ-thio]triphosphate) or ATP[S] (adenosine 5′-[γ-thio]triphosphate) (1 mM solution) into an adiabatic cell containing CENTG1 (100 μM), and the heat energy change accompanying the reaction was detected upon each injection by comparison with a reference cell. Protein solution was placed in the 1.4 ml calorimeter cell and stirred to ensure rapid mixing, and 10 μl aliquots of the nucleotide analogue (titrant) were injected over 10 s with a 5 min interval between each injection until saturation. The titrant injected into buffer alone was used as a negative control. The heat changes were integrated after subtracting values obtained when CENTG1 was titrated into buffer, plotted against the molar ratio of CENTG1/NTP and analysed as a non-linear least-squares fit. Data were analysed with the ORIGIN software package supplied by MicroCal.
RESULTS AND DISCUSSION
Structure of the CENTG1 NTPase domain
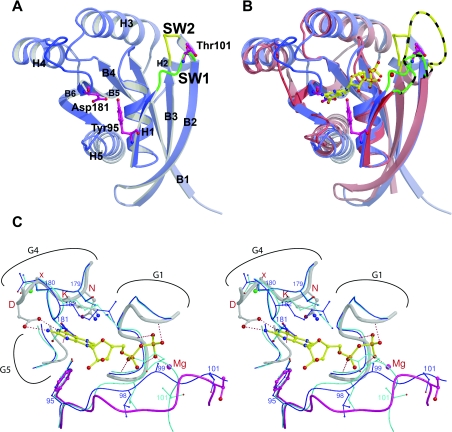
The crystal structure of the GLD (residues 66–241) of CENTG1 was determined in two different space groups, in C2 to a resolution of 2.1 Å (1 Å=0.1 nm) and in P212121 to a resolution of 1.6 Å. The structures were very similar to each other [rmsd (root mean square deviation) of 0.69 Å over 166 Cα atoms], with significant differences only in the region of the loop connecting helix H1 with β-strand B2 (the G2 nucleotide-binding motif also known as Switch I). For both structures, all of the important loops involved in nucleotide binding and hydrolysis were fully resolved. Figure 3(A) shows the overall fold and important residues for the C2 crystal form. In this structure, the Thr101 side chain points away in solution; however, in the in P212121 crystal form, this threonine side chain flips down close to but beneath the expected Mg2+-ion-binding site. Notably, in neither space group was any nucleotide or Mg2+ ion present in the nucleotide-binding site, with the electron density revealing only water molecules where the nucleotide would be expected to bind. In the analysis and discussion below, the C2 space group structure was used unless specified otherwise. A summary of the structure refinement and model statistics is given in Table 1.
Figure 3. CENTG1 GLD structure.
(A) Overview of the CENTG1 GLD structure. Ribbon representation with the secondary-structural elements labelled. The Switch I (SW1) region is in green and the Switch II (SW2) region is in yellow. (B) Superimposition of CENTG1 (blue) GLD and the inactive form of c-H-Ras (PDB code 4Q21) (red). The GDP (in yellow) and Mg2+ ion (red sphere) of c-H-ras are also included. The Switch I and Switch II loops of c-H-ras are coloured as for CENTG1 GLD, with the addition of black stripes. (C) Close-up stereo view of the nucleotide-binding site. Ras is shown in thicker lines, with its Switch I loop in pink, the GDP bound to Ras in yellow and the Mg2+ ion as a pink sphere. The two crystal forms of CENTG1 are shown in dark blue (C2, PDB code 2BMJ) and light blue (P212121, PDB code 2IWR) using thinner lines. Relevant CENTG1 residues are numbered in blue and the NKXD G4 motif of Ras is marked with red letters.
The overall structure of the domain resembles that of other structurally characterized small GTPases, for example c-H-ras (Figure 3B). The protein adopts a classical nucleotide-binding fold consisting of a six-stranded β-sheet surrounded by five α-helices. The five α-helices (H1–H5) and six β-strands (B1–B6) connect with a B1-H1-B2-B3-H2-B4-H3-B5-H4-B6-H5 topology (Figure 1B). Superimposition of CENTG1 with the structure of c-H-ras bound to GDP (PDB code 4Q21) results in an rmsd of 1.26 Å over 135 Cα atoms (Figures 3B and 3C). The GTPase domain of CENTG1 shares moderate sequence identity with the Ras (28% with RAP2A, gi:10518344; 24% with M-ras, gi:32189357; 23% with v-Ha-ras, gi:4885425) and Rab (25% with RAB5A, gi:19923262; 25% with RAB11B, gi:4758986; 25% with Rab9A, gi:4759012) subfamily members of small GTPases, thus rendering the classification of the CENTG1 N-terminal GLD unclear.
There are three regions that show significant differences in their conformation in comparison with benchmark GTPases such as H-ras. They are the Switch I loop, the loop connecting β-strand B3 with H2 (the G3 nucleotide-binding motif or Switch II), and residues Thr179–Arg182 (equivalent to the G4 nucleotide-binding motif, Figure 1B) (Figure 3). These differences can only be partially explained by the lack of a nucleotide in the active site, since there are significant sequence differences in these regions between CENTG1 and typical GTPases.
Active-site structure
In the absence of nucleotide, parts of the protein obscure the volume in which the nucleotide would normally reside. These regions of the protein chain are presumably in different conformations in the presence of bound nucleotide. Figure 3(C) shows the structural differences between CENTG1 GLD and the inactive form of H-ras at the nucleotide-binding site. Included in the superimposition is the location of the GDP molecule present only in the H-ras structure.
In both space groups, the N-terminal part of the Switch I loop has a similar conformation in which Leu98 and Glu99 are positioned such that the backbone carbonyl group of Leu98 occludes the binding pocket for the α-phosphate of a bound nucleotide. The C-terminal part of Switch I is one residue shorter than its equivalent in H-ras or Rab9 (Figure 1B). This part of Switch I occupies a different conformation in each of the structures. It is unlikely that either conformation represents the conformation when bound to GTP or GDP, but it is interesting to note that, in the C2 space group, the side chain of the conserved Thr101 is in a similar position to the equivalent threonine residue (Thr35 in H-ras) in the classical GDP-bound inactive state (Figures 3B and 3C) [5].
The role of this Switch I threonine in binding, hydrolysis and biological function, including effector-mediated interaction, is well established. It plays an important nucleotide-binding role when the GTPase is in the active GTP-bound state, where it co-ordinates the Mg2+ ion with its side chain and the γ-phosphate with its main-chain amide. As such, it is important for interactions with effector proteins, such as in RhoA where the equivalent threonine-to-asparagine mutation prevented activation of phospholipase D [25] and in Ras where a T35A mutant reduced the binding affinity for the Ras-binding domain of the Raf-1 protein kinase by a factor of 188 [26]. The threonine residue is most likely to be invariant in Ras-related proteins, and even T35S mutants show loss of function. For instance, a T35S mutant of Ras showed different dynamic behaviour in its Switch I loop and a more prevalent non-effector-binding conformation [27].
The lack of any bound nucleotide came as a surprise, not least since small GTPases typically have a low-nanomolar affinity for GDP [28], and a number of unusual structural features called into question the ability of this GLD to bind a specific nucleotide. Generally, for the family of small GTPases, the guanine-nucleotide-binding site fits tightly around the ring system, with a number of hydrophobic as well as charged interactions combining to stabilize the nucleotide. The residues that form the classical small GTPase guanine-binding site, as exemplified by H-ras, are the conserved NKXD sequence of the G4 motif [29–31], the phenylalanine side chain immediately C-terminal to H1 on the loop between H1 and B2, and the conserved TSAK sequence of the G5 motif (Figures 1B and 3). All of these residues are highly conserved throughout the family of small GTPases. The NKXD motif interacts with the ring system via the lysine side chain, forming a hydrophobic interaction by running parallel with the ring system plane and, more importantly, the aspartate side chain co-ordinating the two nitrogen groups of the purine (Figure 3C). The aspartate–guanine interaction is primarily responsible for achieving guanine nucleotide specificity. The phenylalanine residue packs at right angles to the ring system plane on the opposite side to the NKXD lysine side chain (Figure 3B), while the conserved residues of the G5 motif interact both with the G4 motif and with the base of the nucleotide.
For CENTG1 GLD, the equivalent of the NKXD motif is TQDR (Figure 1B). In both space groups, the side chain of the aspartate residue from this loop (Asp181) occupies part of the place where the guanine ring would reside (Figure 3C). It is plausible that a rearrangement of this loop could allow a similar interaction between Asp181 and a guanine nucleotide as in typical GTPases; however, the layered interaction equivalent to the NKXD lysine is likely to be lost. Mutational analyses in Ras to determine the role of the Asp181 equivalent in many small GTPases have resulted in a protein that has a very high turnover of GTP and an extremely low affinity for nucleotides [32]. The mutation to alanine resulted in loss of function [33]. The mutation to asparagine resulted in a change of specificity of nucleotide recognition from guanine to xanthine without any loss of hydrolytic function or effector interaction [34]. For CENTG1, a tyrosine residue (Tyr95) replaces the phenylalanine after H1 and the extra polar hydroxy group could also destabilize any bound nucleotide by steric hindrance or electronic interaction (Figure 3C). Only the G5 motif is relatively conserved in CENTG1.
The Switch I and II regions of small GTPases undergo a conformational change depending on whether GDP (inactive state) or GTP (active state) is bound. While the Switch I loop of CENTG1 adopts different conformations in the two crystal forms that we have determined, the Switch II loop adopts the same conformation in both (Figure 3C). Examination of the sequence alignment (Figure 1B) indicates a five-residue deletion with respect to the sequences of H-Ras or Rab9, which results in the shorter Switch II loop in the structure (Figures 3B and 3C). This is likely to have implications for binding to effector proteins, but, in addition, CENTG1 GLD is missing a glutamine residue in this Switch II region (equivalent to Gln61 in Ras), a highly conserved residue in other small GTPases and known to play an important role in GTP hydrolysis [4,28,35]. Gln61 has been shown to play a vital role in GAP-mediated GTP hydrolysis by positioning a water molecule in the transition state aided by the arginine finger contributed by the GAP molecule [35–38]. Interestingly, the tRNA modification GTPase MnmE also lacks this glutamine residue and has relatively high GTPase activity (kcat 10.2 min−1, Km>0.5 mM) [39].
Taken together, these sequence and structural differences prompted us to examine the enzymatic activity and specificity of the CENTG1 GLD.
Nucleotide hydrolysis by CENTG1
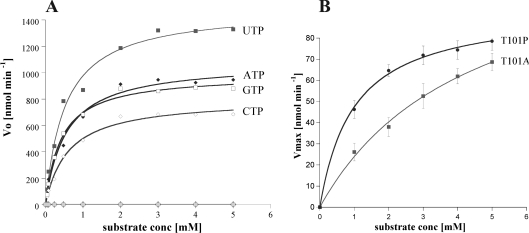
CENTG1 GLD actively catalyses the hydrolysis of GTP, ATP, UTP and CTP (Figure 4A). Multiple turnover conditions at differing substrate concentrations were used to analyse the catalytic properties of the enzyme (Table 2). The kinetic values show that the enzyme is active and the intrinsic nucleotide turnover rate is relatively high compared with typical rates of GTPases. The kcat of CENTG1 with the NTPs is between 16 and 29 min−1 at pH 7.0, depending on the nucleotide (Table 2). Although the rate of hydrolysis is not comparable with that in the presence of GAPs, it is higher than the ‘quick’ turnover of small GTPases such as Rab5 and Ras, for which kcat values of 2×10−1 min−1 and 2.8×10−2 min−1 have been reported [40,41]. The efficient hydrolysis of UTP, ATP and CTP shows that CENTG1 is an NTPase and not simply a GTPase as reported previously [42]. To confirm that the hydrolysis was effected only by CENTG1 and not by a contaminating protein, the GTP and UTP hydrolysis was measured for each of the individual fractions eluted from a gel filtration. This showed clearly that the activity was proportional to the enzyme concentration present in each fraction (Figure 2).
Figure 4. Hydrolysis activity of the wild-type and mutant CENTG1 NTPase domains.
(A) Plot of the initial rate of reaction Vo against concentration of nucleotide triphosphates (mM), fitted to the Michaelis–Menten equation using Kaleidograph software. Hydrolytic rates determined in the absence of enzyme or nucleotides are shown respectively as light grey diamonds and squares along the x-axis. Other lines are labelled. (B) GTP hydrolysis of CENTG1 mutants. Plot of Vmax against substrate concentration (mM) fitted as for wild-type.
Table 2. Kinetic and binding parameters of CENTG1 GLD and mutants.
| Protein | Substrate | pH | Vmax (μM·min−1) | kcat (min−1) | Km (μM) | kcat/Km (μM−1·min−1) |
|---|---|---|---|---|---|---|
| Wild-type | GTP | 7.0 | 2.16 | 21.40 | 541 | 0.039 |
| 8.0 | 0.43 | 4.62 | 442 | 0.010 | ||
| 6.0 | 0.47 | 4.67 | 427 | 0.010 | ||
| Wild-type | ATP | 7.0 | 1.91 | 19.60 | 412 | 0.047 |
| 8.0 | 0.45 | 4.89 | 378 | 0.013 | ||
| 6.0 | 0.48 | 4.53 | 412 | 0.011 | ||
| Wild-type | UTP | 7.0 | 2.81 | 28.62 | 517 | 0.055 |
| 8.0 | 0.65 | 6.73 | 467 | 0.014 | ||
| 6.0 | 0.36 | 3.57 | 449 | 0.007 | ||
| Wild-type | CTP | 7.0 | 1.71 | 16.14 | 601 | 0.026 |
| 8.0 | 0.33 | 3.06 | 516 | 0.006 | ||
| 6.0 | 0.34 | 3.41 | 505 | 0.006 | ||
| T101P | GTP | 7.0 | 0.076 | 0.74 | 990 | |
| 8.0 | 0.012 | 0.12 | 889 | |||
| 6.0 | 0.017 | 0.14 | 901 | |||
| T101A | GTP | 7.0 | 0.089 | 1.08 | 2681 | |
| 8.0 | 0.037 | 0.41 | 4026 | |||
| 6.0 | 0.030 | 0.34 | 3062 |
The CENTG1 GLD showed hydrolytic activity of all the ribonucleotide triphosphates in the pH range analysed (6.0–8.0) and substrate concentrations (Table 2). These observations, combined with the sequence differences discussed above, in particular the absence of the classical NKXD G4 motif, the substitution of phenylalanine for Tyr95 on Switch I and the shorter Switch I loop, suggest the protein should be classified as a member of the P-Loop NTPase superfamily rather than part of the small GTPase family.
Two papers describe the characterization of the full-length and the GLD of centaurins with opposing results. Nie et al. [43] were not able to detect GDP, GTP, ADP or ATP binding or GTPase activity for both the GLD and the full-length mouse CENTG2, which is identical in sequence over the GLD with human CENTG2. On the other hand, Xia et al. [42] were able to measure GTPase activity for the GLD and the full-length protein of human CENTG2 with the GLD enzymatic activity increasing ∼10-fold in the presence of its C-terminal GAP domain. They also recorded a high GDP dissociation rate for the GLD alone. Neither study examined nucleotide selectivity for either the full-length protein or the GLD. From the published papers, it is unclear why, for essentially the same domain, GTPase activity was recorded by Xia et al. [42], but not by Nie et al. [43].
Our results agree more with those of Xia et al. [42] in that our centaurin GLD is active. However, the turnover rates are dramatically different: 0.01 min−1 [42] compared with 21.4 min−1 for GTP (Table 2). We attribute this large difference partly to the different pH values at which the experiments were recorded: pH 8.0 by Xia et al. [42] as compared with our study at pH 7.0, which is closer to the optimal pH for catalytic activity (Table 2), but mostly due to the differences between the two centaurin isoforms: CENTG2 in the work published previously and CENTG1 for this study. Interestingly, the reported 10-fold acceleration in the rate of hydrolysis by CENTG2 due to its GAP domain is unusually low. Typically, GAP domains activate the hydrolysis of small GTPases by a factor of 104–105 [44,45]. The Switch I loop is known to play a central role in the catalytic process of GTPases, as an almost invariant threonine side chain within the loop complexes the Mg2+ ion and interacts with the γ-phosphate group in the active GTP-bound state only. CENTG1 has a threonine residue in the equivalent position (Figures 1B and 3C), whereas CENTG2 has a proline residue, thus making it likely that catalysis for CENTG2 would be different from that of CENTG1.
In order to test this hypothesis, we created T101A and T101P point mutations of CENTG1 to assess the importance of this residue for GTP hydrolysis. Overall, we observed a 30–40-fold reduction in the kcat for T101A and T101P compared with wild-type CENTG1 (Figure 4B, Table 2), illustrating the importance of Thr101 in the hydrolysis mechanism, where, by analogy with Ras, it would be expected to contribute to the octahedral co-ordination of the Mg2+ in the transition state. However, our measured rate of hydrolysis at pH 8.0 for CENTG1 is still an order of magnitude greater than that determined for CENTG2 [42].
Nucleotide affinity of the CENTG1 GLD
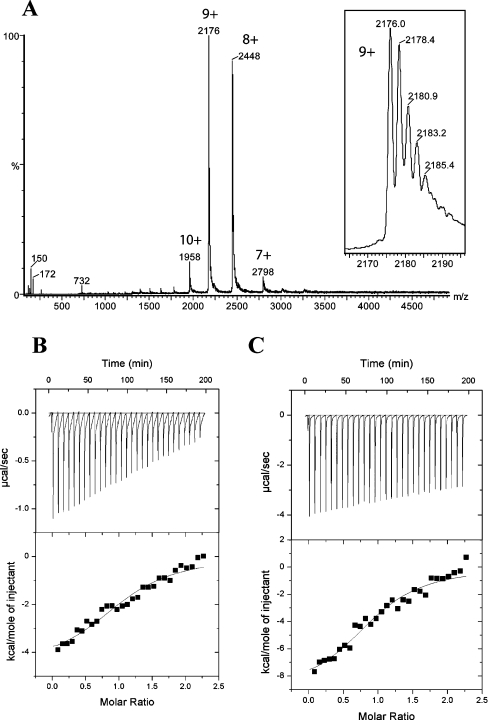
The values of Km of between 400 and 600 μM obtained in the activity measurements (Table 2) are very different from the typical picomolar range of dissociation constants found in Ras family GTPases [41]. Therefore we attempted to confirm nucleotide binding to the CENTG1 NTPase domain by non-covalent MS [46]. ATP, GTP and GDP were screened for binding in the presence and absence of Mg2+. Figure 5(A) shows a nano-electrospray ionization mass spectrum of the CENTG1 NTPase (9 μM) in the presence of a 10-fold excess of GTP. The experiments were performed using sample and instrument parameters which preserve non-covalent complexes intact in the mass spectrometer. The main peak series corresponding to charge states from 7+ to 10+ and a mass of 19575±8 Da (calculated from amino acid sequence: 19581.1 Da) indicates that the CENTG1 NTPase does not have any nucleotide bound under these conditions. No evidence for ATP, GTP or GDP bound to the protein was obtained.
Figure 5. Nucleotide-binding analysis.
Nano-electrospray ionization MS analysis of the CENTG1 NTPase domain in the presence of a 10-fold excess of ribonucleotide triphosphates does not show any nucleotide bound to the protein. The highlighted peaks correspond to charge states of the protein (calculated mass 19581.1 Da, measured mass 19575±8 Da). The inset shows Na+ attachment to the 9+ peak. (B) ITC of interactions between CENTG1 and GTP[S]. The upper panel displays raw energy changes during titration (time), and in the lower panel the derived integrated total energy changes as a function of the molar ratio of the interactants. The yielded thermodynamic parameters are listed in Table 3. (C) ITC interactions between CENTG1 and ATP[S]. The representation is the same as for (B).
We then used ITC to measure the real affinity of CENTG1 for GTP[S] and ATP[S] nucleotide analogues in the presence and absence of Mg2+ ions. Two parameters are measured by this technique, the change in the heat content of the system (enthalpy, ΔH), the tendency being negative, and the change in disorder (entropy, ΔS), the tendency being positive [47]. The best fit was obtained by applying a one-site binding model involving exothermic reaction phases (negative enthalpy changes) with similar favourable free energy changes. The binding assay determined a very weak binding for both the NTP analogues. Analysis of the data indicates that complete saturation of the binding site is not achieved (Figures 5B and 5C). This is likely to be due to the high dissociation rate from the active site. The stoichiometry of CENTG1 binding to these NTP analogues is identical (1:1 ratio), and the association constant Ka is similar, showing no preferential binding to either of the nucleotides (Table 3). The kcat/Km values, which indicate substrate specificity determined from the hydrolytic experiments, also imply no preferential substrate selectivity among the NTPs analysed (Table 2). No binding of CENTG1 with ATP[S] and GTP[S] was measurable in the absence of Mg2+ (results not shown) thus showing that the affinity for NTPs is completely lost in the absence of Mg2+. Assays to measure association constants for CENTG1 with all of the nucleotide diphosphates in the presence or absence of Mg2+ ions did not show any binding, implying extremely low association rates and/or very high dissociation rates. The low affinity for the nucleotides explains the absence of bound nucleotide in the crystal structures, but also highlights the unusual nucleotide-binding properties of CENTG1 in comparison with those of typical small GTPases.
Table 3. Thermodynamic parameters of CENTG1 interaction.
N is the stoichiometry of binding. 1 cal≈4.184 J.
| NTP | N | Ka (M−1) | ΔH (kcal·M−1) | ΔS (cal·M−1·K−1) | ΔG (kcal·M−1) |
|---|---|---|---|---|---|
| GTP[S] | 0.98 | (4.13±1.0)×104 | −4.56±0.3 | 5.56 | −4.56 |
| ATP[S] | 0.98 | (9.33±2.5)×104 | −9.15±0.7 | −8.462 | −9.32 |
Kinetic properties of CENTG1 in comparison with other non-classical GTPases
The high affinity of Ras GTPases for GDP results from a low dissociation rate of the order of 10−5 s−1 [48]. The lower affinity of CENTG1 for nucleotides presumably accounts for its higher turnover rate. The high intrinsic NTPase activity and low affinity for nucleotide di- or tri-phosphates resemble the properties of the large GTPase dynamin, involved in membrane vesicular trafficking, and related enzymes (reviewed in [49]). The GTPase properties of dynamin are distinct from those of classical Ras GTPases [50], with the dissociation rate of GTP from dynamin 104 times higher than from Ras GTPases [51,52]. The kcat of CENTG1 with the different NTPs (16–28 min−1) is comparable with that of wild-type dynamin turnover on lipid tubules (105±47 min−1) [50]. As mentioned above, the atypical GTPase MnmE, which controls the modification of uridine at the wobble position of certain tRNAs [53,54], also exhibits nucleotide activity similar to that of CENTG1 GLD [39].
Conclusions
In human glioblastoma cells, CENTG1 overexpression prevented apoptosis, which was not the case for a dominant-negative mutant of CENTG1 [13]. It was found that the dominant-negative mutant has two mutations (K83A and S84N; Figure 1B) that reside in the P-loop of the NTPase domain. This is consistent with our structural and functional data, as the double mutation will reduce the ability of the NTPase domain to bind the γ-phosphate of GTP and have little or no effect on the binding site for the ring systems. Hence the structure provides an explanation for the inability of the double mutant to bind GTP.
Owing to the relatively high affinity that typical small GTPases generally have for the guanine nucleotide, another protein, functionally known as a GEF, is usually required to aid nucleotide displacement. For CENTG1, the SH3 (Src homology 3) domain of phospholipase Cγ1 has been identified as a GEF [55]. However, as we have shown that the GLD of CENTG1 has a naturally low affinity for the nucleotide triphosphates, the need for a GEF is debatable, which has implications for the cellular action of CENTG1 as it is clearly different from the classical small GTPases in terms of specificity and activity. The smaller Switch II loop (Figure 3B) of CENTG1 as compared with the classical small GTPase family would also be expected to offer a smaller enzyme–GEF interaction surface [56]. Hence, more research is required to uncover the structural and functional roles that known modulatory proteins, such as phospholipase Cγ1 and even CENTG1's own GAP domain, play.
The five signature sequences (G1–G5, Figure 1B) of the small GTPases define the binding site for both the guanine ring system and the phosphate groups. Lack of structural information on other GTPases or GTPase-like proteins in their apo form and bound to alternative nucleotides prevents a comprehensive explanation of the mechanisms for NTP binding and hydrolysis by CENTG1, but this study provides important information towards that explanation. The CENTG1 NTPase structure has revealed the residues important for nucleotide specificity and, along with enzymatic assays, has demonstrated that this domain should be classified not as a GTPase but as an NTPase.
It therefore appears that CENTG1 is not a classical small GTPase, and in its catalytic properties is related to some atypical large GTPases, exhibiting a rate of hydrolysis as if in the presence of a GEF, with a correspondingly low nucleotide-binding affinity. The unusual nature of this enzyme is confirmed by its broad nucleotide hydrolysis efficiency which is likely to be the result of its unusual G4 motif. These results point to an alternative control mechanism for this enzyme, opening up new avenues of research for this family of proteins.
Acknowledgments
We thank Neil Oldham (Chemistry Department, University of Oxford) for access to the mass spectrometer. The Structural Genomics Consortium is a registered charity (number 1097737) funded by the Wellcome Trust, GlaxoSmithKline, Genome Canada, the Canadian Institutes of Health Research, the Ontario Innovation Trust, the Ontario Research and Development Challenge Fund, the Canadian Foundation for Innovation, VINNOVA (Swedish Governmental Agency for Innovation Systems), The Knut and Alice Wallenberg Foundation, The Swedish Foundation for Strategic Research and Karolinska Institutet.
References
- 1.Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 2.Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 3.Boguski M. S., McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 4.Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milburn M. V., Tong L., deVos A. M., Brunger A., Yamaizumi Z., Nishimura S., Kim S. H. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 6.Schlichting I., Almo S. C., Rapp G., Wilson K., Petratos K., Lentfer A., Wittinghofer A., Kabsch W., Pai E. F., Petsko G. A., et al. Time-resolved X-ray crystallographic study of the conformational change in Ha-Ras p21 protein on GTP hydrolysis. Nature. 1990;345:309–315. doi: 10.1038/345309a0. [DOI] [PubMed] [Google Scholar]
- 7.Coleman D. E., Berghuis A. M., Lee E., Linder M. E., Gilman A. G., Sprang S. R. Structures of active conformations of Giα1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 8.Sondek J., Lambright D. G., Noel J. P., Hamm H. E., Sigler P. B. GTPase mechanism of G proteins from the 1.7-Å crystal structure of transducin α-GDP–AlF4−. Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 9.Colicelli J. Human RAS superfamily proteins and related GTPases. Science STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klarlund J. K., Rameh L. E., Cantley L. C., Buxton J. M., Holik J. J., Sakelis C., Patki V., Corvera S., Czech M. P. Regulation of GRP1-catalyzed ADP ribosylation factor guanine nucleotide exchange by phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:1859–1862. doi: 10.1074/jbc.273.4.1859. [DOI] [PubMed] [Google Scholar]
- 11.Hammonds-Odie L. P., Jackson T. R., Profit A. A., Blader I. J., Turck C. W., Prestwich G. D., Theibert A. B. Identification and cloning of centaurin-α: a novel phosphatidylinositol 3,4,5-trisphosphate-binding protein from rat brain. J. Biol. Chem. 1996;271:18859–18868. doi: 10.1074/jbc.271.31.18859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye K., Hurt K. J., Wu F. Y., Fang M., Luo H. R., Hong J. J., Blackshaw S., Ferris C. D., Snyder S. H. Pike: a nuclear GTPase that enhances PI3kinase activity and is regulated by protein 4.1N. Cell. 2000;103:919–930. doi: 10.1016/s0092-8674(00)00195-1. [DOI] [PubMed] [Google Scholar]
- 13.Ahn J. Y., Hu Y., Kroll T. G., Allard P., Ye K. PIKE-A is amplified in human cancers and prevents apoptosis by up-regulating Akt. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6993–6998. doi: 10.1073/pnas.0400921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du K., Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J. Biol. Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 15.Knobbe C. B., Trampe-Kieslich A., Reifenberger G. Genetic alteration and expression of the phosphoinositol-3-kinase/Akt pathway genes PIK3CA and PIKE in human glioblastomas. Neuropathol. Appl. Neurobiol. 2005;31:486–490. doi: 10.1111/j.1365-2990.2005.00660.x. [DOI] [PubMed] [Google Scholar]
- 16.Ahn J. Y., Rong R., Liu X., Ye K. PIKE/nuclear PI 3-kinase signaling mediates the antiapoptotic actions of NGF in the nucleus. EMBO J. 2004;23:3995–4006. doi: 10.1038/sj.emboj.7600392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leslie A. G. Integration of macromolecular diffraction data. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 19.Storoni L. C., McCoy A. J., Read R. J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 20.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A Found. Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 23.Sobott F., Robinson C. V. Protein complexes gain momentum. Curr. Opin. Struct. Biol. 2002;12:729–734. doi: 10.1016/s0959-440x(02)00400-1. [DOI] [PubMed] [Google Scholar]
- 24.Keetch C. A., Hernanndez H., Sterling A., Baumert M., Allen M. H., Robinson C. V. Use of a microchip device coupled with mass spectrometry for ligand screening of a multi-protein target. Anal. Chem. 2003;75:4937–4941. doi: 10.1021/ac034251c. [DOI] [PubMed] [Google Scholar]
- 25.Bae C. D., Min D. S., Fleming I. N., Exton J. H. Determination of interaction sites on the small G protein RhoA for phospholipase D. J. Biol. Chem. 1998;273:11596–11604. doi: 10.1074/jbc.273.19.11596. [DOI] [PubMed] [Google Scholar]
- 26.Herrmann C., Martin G. A., Wittinghofer A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J. Biol. Chem. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 27.Spoerner M., Herrmann C., Vetter I. R., Kalbitzer H. R., Wittinghofer A. Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4944–4949. doi: 10.1073/pnas.081441398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pai E. F., Kabsch W., Krengel U., Holmes K. C., John J., Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341:209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- 29.Denker B. M., Boutin P. M., Neer E. J. Interactions between the amino- and carboxyl-terminal regions of Gα subunits: analysis of mutated Gαo/Gαi2 chimeras. Biochemistry. 1995;34:5544–5553. doi: 10.1021/bi00016a028. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt G., Lenzen C., Simon I., Deuter R., Cool R. H., Goody R. S., Wittinghofer A. Biochemical and biological consequences of changing the specificity of p21ras from guanosine to xanthosine nucleotides. Oncogene. 1996;12:87–96. [PubMed] [Google Scholar]
- 31.Weijland A., Parlato G., Parmeggiani A. Elongation factor Tu D138N, a mutant with modified substrate specificity, as a tool to study energy consumption in protein biosynthesis. Biochemistry. 1994;33:10711–10717. [Google Scholar]
- 32.Cool R. H., Schmidt G., Lenzen C. U., Prinz H., Vogt D., Wittinghofer A. The Ras mutant D119N is both dominant negative and activated. Mol. Cell. Biol. 1999;19:6297–6305. doi: 10.1128/mcb.19.9.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sigal I. S., Gibbs J. B., D'Alonzo J. S., Temeles G. L., Wolanski B. S., Socher S. H., Scolnik E. M. Mutant ras-encoded proteins with altered nucleotide binding exert dominant biological effects. Proc. Natl. Acad. Sci. U.S.A. 1986;83:952–956. doi: 10.1073/pnas.83.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong J. M., Chen-Hwang M. C., Hwang Y. W. Switching nucleotide specificity of Ha-Ras p21 by a single amino acid substitution at aspartate 119. J. Biol. Chem. 1995;270:10002–10007. doi: 10.1074/jbc.270.17.10002. [DOI] [PubMed] [Google Scholar]
- 35.Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmuller L., Lautwein A., Schmitz F., Wittinghofer A. The Ras–RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 36.Nassar N., Hoffman G. R., Manor D., Clardy J. C., Cerione R. A. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat. Struct. Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- 37.Rittinger K., Walker P. A., Eccleston J. F., Nurmahomed K., Owen D., Laue E., Gamblin S. J., Smerdon S. J. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- 38.Rittinger K., Walker P. A., Eccleston J. F., Smerdon S. J., Gamblin S. J. Structure at 1.65 Å of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 39.Yim L., Martinez-Vicente M., Villarroya M., Aguado C., Knecht E., Armengod M. E. The GTPase activity and C-terminal cysteine of the Escherichia coli MnmE protein are essential for its tRNA modifying function. J. Biol. Chem. 2003;278:28378–28387. doi: 10.1074/jbc.M301381200. [DOI] [PubMed] [Google Scholar]
- 40.John J., Schlichting I., Schiltz E., Rosch P., Wittinghofer A. C-terminal truncation of p21H preserves crucial kinetic and structural properties. J. Biol. Chem. 1989;264:13086–13092. [PubMed] [Google Scholar]
- 41.Hoffenberg S., Sanford J. C., Liu S., Daniel D. S., Tuvin M., Knoll B. J., Wessling-Resnick M., Dickey B. F. Biochemical and functional characterization of a recombinant GTPase, Rab5, and two of its mutants. J. Biol. Chem. 1995;270:5048–5056. doi: 10.1074/jbc.270.10.5048. [DOI] [PubMed] [Google Scholar]
- 42.Xia C., Ma W., Stafford L. J., Liu C., Gong L., Martin J. F., Liu M. GGAPs, a new family of bifunctional GTP-binding and GTPase-activating proteins. Mol. Cell. Biol. 2003;23:2476–2488. doi: 10.1128/MCB.23.7.2476-2488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nie Z., Stanley K. T., Stauffer S., Jacques K. M., Hirsch D. S., Takei J., Randazzo P. A. AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J. Biol. Chem. 2002;277:48965–48975. doi: 10.1074/jbc.M202969200. [DOI] [PubMed] [Google Scholar]
- 44.Lancaster C. A., Taylor-Harris P. M., Self A. J., Brill S., van Erp H. E., Hall A. Characterization of rhoGAP. A GTPase-activating protein for rho-related small GTPases. J. Biol. Chem. 1994;269:1137–1142. [PubMed] [Google Scholar]
- 45.Lamarche N., Hall A. GAPs for rho-related GTPases. Trends Genet. 1994;10:436–440. doi: 10.1016/0168-9525(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 46.Ganguly A. K., Pramanik B. N., Huang E. C., Liberles S., Heimark L., Liu Y. H., Tsarbopoulos A., Doll R. J., Taveras A. G., Remiszewski S., et al. Detection and structural characterization of ras oncoprotein-inhibitors complexes by electrospray mass spectrometry. Bioorg. Med. Chem. 1997;5:817–820. doi: 10.1016/s0968-0896(97)00021-7. [DOI] [PubMed] [Google Scholar]
- 47.Cooper A. Thermodynamic analysis of biomolecular interactions. Curr. Opin. Chem. Biol. 1999;3:557–563. doi: 10.1016/s1367-5931(99)00008-3. [DOI] [PubMed] [Google Scholar]
- 48.John J., Sohmen R., Feuerstein J., Linke R., Wittinghofer A., Goody R. S. Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry. 1990;29:6058–6065. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- 49.Danino D., Hinshaw J. E. Dynamin family of mechanoenzymes. Curr. Opin. Cell Biol. 2001;13:454–460. doi: 10.1016/s0955-0674(00)00236-2. [DOI] [PubMed] [Google Scholar]
- 50.Song B. D., Leonard M., Schmid S. L. Dynamin GTPase domain mutants that differentially affect GTP binding, GTP hydrolysis, and clathrin-mediated endocytosis. J. Biol. Chem. 2004;279:40431–40436. doi: 10.1074/jbc.M407007200. [DOI] [PubMed] [Google Scholar]
- 51.Binns D. D., Helms M. K., Barylko B., Davis C. T., Jameson D. M., Albanesi J. P., Eccleston J. F. The mechanism of GTP hydrolysis by dynamin II: a transient kinetic study. Biochemistry. 2000;39:7188–7196. doi: 10.1021/bi000033r. [DOI] [PubMed] [Google Scholar]
- 52.Neal S. E., Eccleston J. F., Hall A., Webb M. R. Kinetic analysis of the hydrolysis of GTP by p21N-ras: the basal GTPase mechanism. J. Biol. Chem. 1988;263:19718–19722. [PubMed] [Google Scholar]
- 53.Cabedo H., Macian F., Villarroya M., Escudero J. C., Martinez-Vicente M., Knecht E., Armengod M. E. The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J. 1999;18:7063–7076. doi: 10.1093/emboj/18.24.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kruger M. K., Sorensen M. A. Aminoacylation of hypomodified tRNAGlu in vivo. J. Mol. Biol. 1998;284:609–620. doi: 10.1006/jmbi.1998.2197. [DOI] [PubMed] [Google Scholar]
- 55.Ye K., Aghdasi B., Luo H. R., Moriarity J. L., Wu F. Y., Hong J. J., Hurt K. J., Bae S. S., Suh P. G., Snyder S. H. Phospholipase Cγ1 is a physiological guanine nucleotide exchange factor for the nuclear GTPase PIKE. Nature. 2002;415:541–544. doi: 10.1038/415541a. [DOI] [PubMed] [Google Scholar]
- 56.Cherfils J., Chardin P. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]